- Record: found
- Abstract: found
- Article: found
Nucleic acid-based approaches to investigate microbial-related cheese quality defects
Read this article at
Abstract
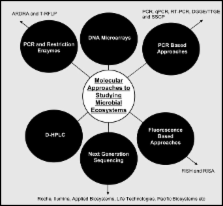
The microbial profile of cheese is a primary determinant of cheese quality. Microorganisms can contribute to aroma and taste defects, form biogenic amines, cause gas and secondary fermentation defects, and can contribute to cheese pinking and mineral deposition issues. These defects may be as a result of seasonality and the variability in the composition of the milk supplied, variations in cheese processing parameters, as well as the nature and number of the non-starter microorganisms which come from the milk or other environmental sources. Such defects can be responsible for production and product recall costs and thus represent a significant economic burden for the dairy industry worldwide. Traditional non-molecular approaches are often considered biased and have inherently slow turnaround times. Molecular techniques can provide early and rapid detection of defects that result from the presence of specific spoilage microbes and, ultimately, assist in enhancing cheese quality and reducing costs. Here we review the DNA-based methods that are available to detect/quantify spoilage bacteria, and relevant metabolic pathways in cheeses and, in the process, highlight how these strategies can be employed to improve cheese quality and reduce the associated economic burden on cheese processors.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
Biogenic amines: their importance in foods.
- Record: found
- Abstract: found
- Article: not found