- Record: found
- Abstract: found
- Article: found
DNA Methylation Clocks and Their Predictive Capacity for Aging Phenotypes and Healthspan
Read this article at
Abstract
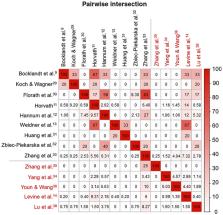
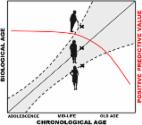
The number of age predictors based on DNA methylation (DNAm) profile is rising due to their potential in predicting healthspan and application in age-related illnesses, such as neurodegenerative diseases. The cumulative assessment of DNAm levels at age-related CpGs (DNAm clock) may reflect biological aging. Such DNAm clocks have been developed using various training models and could mirror different aspects of disease/aging mechanisms. Hence, evaluating several DNAm clocks together may be the most effective strategy in capturing the complexity of the aging process. However, various confounders may influence the outcome of these age predictors, including genetic and environmental factors, as well as technical differences in the selected DNAm arrays. These factors should be taken into consideration when interpreting DNAm clock predictions. In the current review, we discuss 15 reported DNAm clocks with consideration for their utility in investigating neurodegenerative diseases and suggest research directions towards developing a more optimal measure for biological aging.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
DAVID: Database for Annotation, Visualization, and Integrated Discovery.

- Record: found
- Abstract: found
- Article: found
An epigenetic biomarker of aging for lifespan and healthspan
- Record: found
- Abstract: found
- Article: found