- Record: found
- Abstract: found
- Article: found
Non-canonical amino acid labeling in proteomics and biotechnology
Read this article at
Abstract
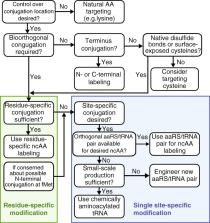
Metabolic labeling of proteins with non-canonical amino acids (ncAAs) provides unique bioorthogonal chemical groups during de novo synthesis by taking advantage of both endogenous and heterologous protein synthesis machineries. Labeled proteins can then be selectively conjugated to fluorophores, affinity reagents, peptides, polymers, nanoparticles or surfaces for a wide variety of downstream applications in proteomics and biotechnology. In this review, we focus on techniques in which proteins are residue- and site-specifically labeled with ncAAs containing bioorthogonal handles. These ncAA-labeled proteins are: readily enriched from cells and tissues for identification via mass spectrometry-based proteomic analysis; selectively purified for downstream biotechnology applications; or labeled with fluorophores for in situ analysis. To facilitate the wider use of these techniques, we provide decision trees to help guide the design of future experiments. It is expected that the use of ncAA labeling will continue to expand into new application areas where spatial and temporal analysis of proteome dynamics and engineering new chemistries and new function into proteins are desired.
Related collections
Most cited references114
- Record: found
- Abstract: found
- Article: not found
Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics.
- Record: found
- Abstract: found
- Article: not found
Cell surface engineering by a modified Staudinger reaction.
- Record: found
- Abstract: found
- Article: not found