- Record: found
- Abstract: found
- Article: found
Myogenin is an essential regulator of adult myofibre growth and muscle stem cell homeostasis
Read this article at
Abstract
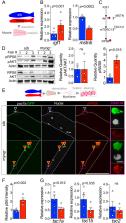
Growth and maintenance of skeletal muscle fibres depend on coordinated activation and return to quiescence of resident muscle stem cells (MuSCs). The transcription factor Myogenin (Myog) regulates myocyte fusion during development, but its role in adult myogenesis remains unclear. In contrast to mice, myog -/- zebrafish are viable, but have hypotrophic muscles. By isolating adult myofibres with associated MuSCs, we found that myog -/- myofibres have severely reduced nuclear number, but increased myonuclear domain size. Expression of fusogenic genes is decreased, Pax7 upregulated, MuSCs are fivefold more numerous and mis-positioned throughout the length of myog -/- myofibres instead of localising at myofibre ends as in wild-type. Loss of Myog dysregulates mTORC1 signalling, resulting in an ‘alerted’ state of MuSCs, which display precocious activation and faster cell cycle entry ex vivo, concomitant with myod upregulation. Thus, beyond controlling myocyte fusion, Myog influences the MuSC:niche relationship, demonstrating a multi-level contribution to muscle homeostasis throughout life.
Related collections
Most cited references103
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.

- Record: found
- Abstract: found
- Article: found
An Integrated Encyclopedia of DNA Elements in the Human Genome
- Record: found
- Abstract: not found
- Article: not found