- Record: found
- Abstract: found
- Article: found
Distinct microbial communities degrade cellulose diacetate bioplastics in the coastal ocean
Read this article at
ABSTRACT
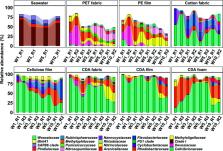
Cellulose diacetate (CDA) is a bio-based plastic widely used in consumer products. CDA is a promising alternative to conventional thermoplastics due to its susceptibility to biodegradation in various environments. Despite widespread evidence for the degradation of CDA, relatively little is known about the microorganisms that drive degradation, particularly in the ocean. Recently, we documented the biodegradation of CDA-based materials (i.e., fabric, film, and foam) in a continuous-flow natural seawater mesocosm on the timescales of months, as indicated by mass loss, enzyme activity, and respiration to carbon dioxide. These findings paved the way for the present study aimed at identifying key microbial taxa implicated in CDA degradation. Analysis based on 16S rRNA gene amplicon sequencing of bacteria and archaea revealed that material type, incubation time, material morphology (e.g., fabric vs film), and plasticizer content significantly influenced the microbial community structure. Differential abundance analysis revealed that bacterial taxa affiliated with the families of Arenicellaceae, Cellvibrionaceae, Methyloligellaceae, Micavibrionaceae, Puniceicoccaceae, Spirosomaceae, and Thermoanaerobaculaceae, and the order of Pseudomonadales potentially initiated the degradation (i.e., deacetylation) of CDA fabric and film. These taxa were notably distinct from CDA-degrading microbes reported in non-seawater environments. Collectively, the findings lend further support for CDA as a promising next-generation, high-utility, and low-environmental persistence bioplastic material.
IMPORTANCE
Cellulose diacetate (CDA) is a promising alternative to conventional plastics due to its versatility in manufacturing and low environmental persistence. Previously, our group demonstrated that CDA is susceptible to biodegradation in the ocean on timescales of months. In this study, we report the composition of microorganisms driving CDA degradation in the coastal ocean. We found that the coastal ocean harbors distinct bacterial taxa implicated in CDA degradation and these taxa have not been previously identified in prior CDA degradation studies, indicating an unexplored diversity of CDA-degrading bacteria in the ocean. Moreover, the shape of the plastic article (e.g., a fabric, film, or foam) and plasticizer in the plastic matrix selected for different microbial communities. Our findings pave the way for future studies to identify the specific species and enzymes that drive CDA degradation in the marine environment, ultimately yielding a more predictive understanding of CDA biodegradation across space and time.
Related collections
Most cited references69

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data

- Record: found
- Abstract: found
- Article: found