- Record: found
- Abstract: found
- Article: found
Approximate Bayesian Computation
Read this article at
Abstract
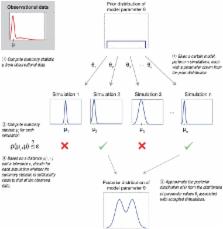
Approximate Bayesian computation (ABC) constitutes a class of computational methods rooted in Bayesian statistics. In all model-based statistical inference, the likelihood function is of central importance, since it expresses the probability of the observed data under a particular statistical model, and thus quantifies the support data lend to particular values of parameters and to choices among different models. For simple models, an analytical formula for the likelihood function can typically be derived. However, for more complex models, an analytical formula might be elusive or the likelihood function might be computationally very costly to evaluate. ABC methods bypass the evaluation of the likelihood function. In this way, ABC methods widen the realm of models for which statistical inference can be considered. ABC methods are mathematically well-founded, but they inevitably make assumptions and approximations whose impact needs to be carefully assessed. Furthermore, the wider application domain of ABC exacerbates the challenges of parameter estimation and model selection. ABC has rapidly gained popularity over the last years and in particular for the analysis of complex problems arising in biological sciences (e.g., in population genetics, ecology, epidemiology, and systems biology).
Related collections
Most cited references45
- Record: found
- Abstract: not found
- Article: not found
Approximate Bayesian Computation in Evolution and Ecology
- Record: found
- Abstract: found
- Article: not found