- Record: found
- Abstract: found
- Article: found
Population genomic analyses of early-phase Atlantic Salmon ( Salmo salar) domestication/captive breeding
Read this article at
Abstract
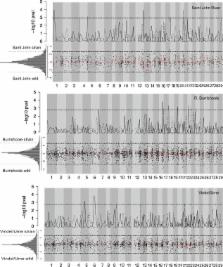
Domestication can have adverse genetic consequences, which may reduce the fitness of individuals once released back into the wild. Many wild Atlantic salmon ( Salmo salarL.) populations are threatened by anthropogenic influences, and they are supplemented with captively bred fish. The Atlantic salmon is also widely used in selective breeding programs to increase the mean trait values for desired phenotypic traits. We analyzed a genomewide set of SNPs in three domesticated Atlantic salmon strains and their wild conspecifics to identify loci underlying domestication. The genetic differentiation between domesticated strains and wild populations was low ( F ST < 0.03), and domesticated strains harbored similar levels of genetic diversity compared to their wild conspecifics. Only a few loci showed footprints of selection, and these loci were located in different linkage groups among the different wild population/hatchery strain comparisons. Simulated scenarios indicated that differentiation in quantitative trait loci exceeded that in neutral markers during the early phases of divergence only when the difference in the phenotypic optimum between populations was large. This study indicates that detecting selection using standard approaches in the early phases of domestication might be challenging unless selection is strong and the traits under selection show simple inheritance patterns.
Related collections
Most cited references76
- Record: found
- Abstract: found
- Article: not found
Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques.
- Record: found
- Abstract: found
- Article: not found
Molecular signatures of natural selection.
- Record: found
- Abstract: found
- Article: not found