- Record: found
- Abstract: found
- Article: found
The LAMMER Kinase Homolog, Lkh1, Regulates Tup Transcriptional Repressors through Phosphorylation in Schizosaccharomyces pombe*
Read this article at
Abstract
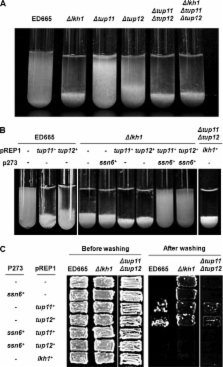
Disruption of the fission yeast LAMMER kinase, Lkh1, gene resulted in diverse phenotypes, including adhesive filamentous growth and oxidative stress sensitivity, but an exact cellular function had not been assigned to Lkh1. Through an in vitro pull-down approach, a transcriptional repressor, Tup12, was identified as an Lkh1 binding partner. Interactions between Lkh1 and Tup11 or Tup12 were confirmed by in vitro and in vivo binding assays. Tup proteins were phosphorylated by Lkh1 in a LAMMER motif-dependent manner. The LAMMER motif was also necessary for substrate recognition in vitro and cellular function in vivo. Transcriptional activity assays using promoters negatively regulated by Tup11 and Tup12 showed 6 or 2 times higher activity in the Δ lkh1 mutant than the wild type, respectively. Northern analysis revealed derepressed expression of the fbp1 + mRNA in Δ lkh1 and in Δ tup11Δ tup12 mutant cells under repressed conditions. Δ lkh1 and Δ tup11Δ tup12 mutant cells showed flocculation, which was reversed by co-expression of Tup11 and -12 with Ssn6. Here, we presented a new aspect of the LAMMER kinase by demonstrating that the activities of global transcriptional repressors, Tup11 and Tup12, were positively regulated by Lkh1-mediated phosphorylation.
Related collections
Most cited references58
- Record: found
- Abstract: found
- Article: not found
A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae.
- Record: found
- Abstract: found
- Article: not found
Rapid and efficient site-specific mutagenesis without phenotypic selection.
- Record: found
- Abstract: found
- Article: not found