- Record: found
- Abstract: found
- Article: not found
Staphylococcus aureus host interactions and adaptation
Read this article at
Abstract
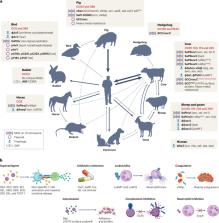
Invasive Staphylococcus aureus infections are common, causing high mortality, compounded by the propensity of the bacterium to develop drug resistance. S. aureus is an excellent case study of the potential for a bacterium to be commensal, colonizing, latent or disease-causing; these states defined by the interplay between S. aureus and host. This interplay is multidimensional and evolving, exemplified by the spread of S. aureus between humans and other animal reservoirs and the lack of success in vaccine development. In this Review, we examine recent advances in understanding the S. aureus–host interactions that lead to infections. We revisit the primary role of neutrophils in controlling infection, summarizing the discovery of new immune evasion molecules and the discovery of new functions ascribed to well-known virulence factors. We explore the intriguing intersection of bacterial and host metabolism, where crosstalk in both directions can influence immune responses and infection outcomes. This Review also assesses the surprising genomic plasticity of S. aureus, its dualism as a multi-mammalian species commensal and opportunistic pathogen and our developing understanding of the roles of other bacteria in shaping S. aureus colonization.
Abstract
In this Review, Howden and co-workers examine and integrate recent key advances in understanding the mechanisms that Staphylococcus aureus uses to cause infections.
Related collections
Most cited references177
- Record: found
- Abstract: found
- Article: not found
Myeloid-derived suppressor cells as regulators of the immune system.
- Record: found
- Abstract: found
- Article: not found
Succinate is an inflammatory signal that induces IL-1β through HIF-1α.
- Record: found
- Abstract: found
- Article: not found
