- Record: found
- Abstract: found
- Article: found
The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis

Read this article at
Abstract
The antagonistic crosstalk between gibberellic acid (GA) and abscisic acid (ABA) plays a pivotal role in the modulation of seed germination. However, the molecular mechanism of such phytohormone interaction remains largely elusive. Here we show that three Arabidopsis NUCLEAR FACTOR-Y C (NF-YC) homologues NF-YC3, NF-YC4 and NF-YC9 redundantly modulate GA- and ABA-mediated seed germination. These NF-YCs interact with the DELLA protein RGL2, a key repressor of GA signalling. The NF-YC–RGL2 module targets ABI5, a gene encoding a core component of ABA signalling, via specific CCAAT elements and collectively regulates a set of GA- and ABA-responsive genes, thus controlling germination. These results suggest that the NF-YC–RGL2–ABI5 module integrates GA and ABA signalling pathways during seed germination.
Abstract
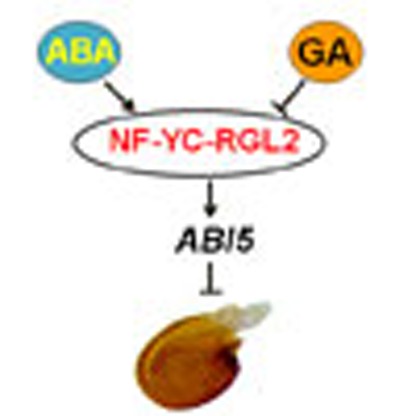 Crosstalk between gibberellic acid (GA) and abscisic acid (ABA) regulates seed germination.
Here the authors show that NF-YC transcription factors can interact with the RGL2
DELLA protein to regulate expression of
ABI5 and therefore modulate ABA- and GA-responsive gene expression.
Crosstalk between gibberellic acid (GA) and abscisic acid (ABA) regulates seed germination.
Here the authors show that NF-YC transcription factors can interact with the RGL2
DELLA protein to regulate expression of
ABI5 and therefore modulate ABA- and GA-responsive gene expression.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Seed germination and vigor.
- Record: found
- Abstract: found
- Article: not found