- Record: found
- Abstract: found
- Article: found
BK Polyomavirus Evades Innate Immune Sensing by Disrupting the Mitochondrial Network and Promotes Mitophagy

Read this article at
Summary
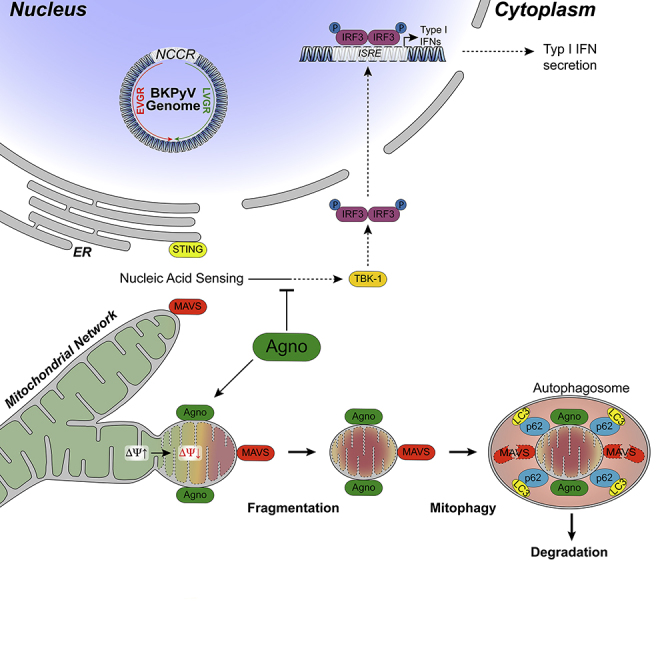
Immune escape contributes to viral persistence, yet little is known about human polyomaviruses. BK-polyomavirus (BKPyV) asymptomatically infects 90% of humans but causes premature allograft failure in kidney transplant patients. Despite virus-specific T cells and neutralizing antibodies, BKPyV persists in kidneys and evades immune control as evidenced by urinary shedding in immunocompetent individuals. Here, we report that BKPyV disrupts the mitochondrial network and membrane potential when expressing the 66aa-long agnoprotein during late replication. Agnoprotein is necessary and sufficient, using its amino-terminal and central domain for mitochondrial targeting and network disruption, respectively. Agnoprotein impairs nuclear IRF3-translocation, interferon-beta expression, and promotes p62/SQSTM1-mitophagy. Agnoprotein-mutant viruses unable to disrupt mitochondria show reduced replication and increased interferon-beta expression but can be rescued by type-I interferon blockade, TBK1-inhibition, or CoCl 2-treatment. Mitochondrial fragmentation and p62/SQSTM1-autophagy occur in allograft biopsies of kidney transplant patients with BKPyV nephropathy. JCPyV and SV40 infection similarly disrupt mitochondrial networks, indicating a conserved mechanism facilitating polyomavirus persistence and post-transplant disease.
Graphical Abstract
Highlights
-
•
BK polyomavirus agnoprotein disrupts mitochondrial membrane potential and network
-
•
Agnoprotein impairs nucleus IRF3 translocation and interferon-β expression
-
•
Agnoprotein facilitates innate immune evasion during the late viral replication phase
-
•
Damaged mitochondria are targeted for p62/SQSTM1 autophagy
Abstract
Biological Sciences; Immunology; Virology; Cell Biology
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity.
- Record: found
- Abstract: found
- Article: not found
Selective autophagy mediated by autophagic adapter proteins.
- Record: found
- Abstract: found
- Article: not found