- Record: found
- Abstract: found
- Article: found
Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
Read this article at
Abstract
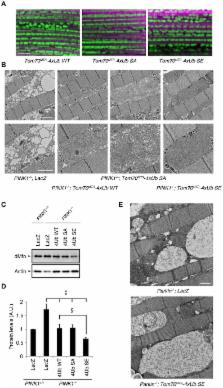
The kinase PINK1 and the E3 ubiquitin (Ub) ligase Parkin participate in mitochondrial quality control. The phosphorylation of Ser65 in Parkin's ubiquitin-like (UBl) domain by PINK1 stimulates Parkin activation and translocation to damaged mitochondria, which induces mitophagy generating polyUb chain. However, Parkin Ser65 phosphorylation is insufficient for Parkin mitochondrial translocation. Here we report that Ser65 in polyUb chain is also phosphorylated by PINK1, and that phosphorylated polyUb chain on mitochondria tethers Parkin at mitochondria. The expression of Tom70 MTS-4xUb SE, which mimics phospho-Ser65 polyUb chains on the mitochondria, activated Parkin E3 activity and its mitochondrial translocation. An E3-dead form of Parkin translocated to mitochondria with reduced membrane potential in the presence of Tom70 MTS-4xUb SE, whereas non-phospho-polyUb mutant Tom70 MTS-4xUb SA abrogated Parkin translocation. Parkin binds to the phospho-polyUb chain through its RING1-In-Between-RING (IBR) domains, but its RING0-linker is also required for mitochondrial translocation. Moreover, the expression of Tom70 MTS-4xUb SE improved mitochondrial degeneration in PINK1-deficient, but not Parkin-deficient, Drosophila. Our study suggests that the phosphorylation of mitochondrial polyUb by PINK1 is implicated in both Parkin activation and mitochondrial translocation, predicting a chain reaction mechanism of mitochondrial phospho-polyUb production by which rapid translocation of Parkin is achieved.
Author Summary
Parkinson's disease is a neurodegenerative disorder caused by degeneration of the midbrain dopaminergic system in addition to other nervous systems. PINK1 and parkin, which encode mitochondrial protein kinase and cytosolic Ub ligase, respectively, were identified as the genes responsible for the autosomal recessive form of juvenile Parkinson's disease. Activation of PINK1 upon reduction of mitochondrial membrane potential recruits Parkin from the cytosol activating its Ub ligase activity, which ensures removal of damaged mitochondria through mitophagy. However, how PINK1 recruits Parkin to the damaged mitochondria remained unclear. Here, we describe that the phosphorylation of polyUb chain by PINK1 is a key event to recruit Parkin on the mitochondria. Parkin binds to, and is activated by, phospho-polyUb generated by Parkin in collaboration with PINK1. Expression of a phospho-polyUb mimetic protein on mitochondria rescued mitochondrial degeneration caused by loss of PINK1 in Drosophila. Our study suggests the existence of an amplification cascade of Parkin activation and mitochondrial translocation, in which a ‘seed' of phosphorylated polyUb on the mitochondria, generated by PINK1 and Parkin, triggers a chain reaction of Parkin recruitment and activation.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
A probability-based approach for high-throughput protein phosphorylation analysis and site localization.

- Record: found
- Abstract: found
- Article: found
PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65
- Record: found
- Abstract: found
- Article: not found