- Record: found
- Abstract: found
- Article: found
In silico designing of multiepitope-based-peptide (MBP) vaccine against MAPK protein express for Alzheimer's disease in Zebrafish

Read this article at
Abstract
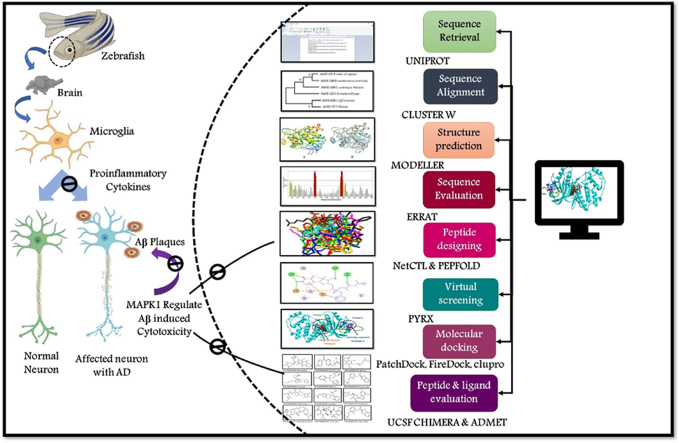
Understanding the role of the mitogen-activated protein kinases (MAPKs) signalling pathway is essential in advancing treatments for neurodegenerative disorders like Alzheimer's. In this study, we investigate in-silico techniques involving computer-based methods to extract the MAPK1 sequence. Our applied methods enable us to analyze the protein's structure, evaluate its properties, establish its evolutionary relationships, and assess its prevalence in populations. We also predict epitopes, assess their ability to trigger immune responses, and check for allergenicity using advanced computational tools to understand their immunological properties comprehensively. We apply virtual screening, docking, and structure modelling to identify promising drug candidates, analyze their interactions, and enhance drug design processes. We identified a total of 30 cell-targeting molecules against the MAPK1 protein, where we selected top 10 CTL epitopes (PAGGGPNPG, GGGPNPGSG, SAPAGGGPN, AVSAPAGGG, AGGGPNPGS, ATAAVSAPA, TAAVSAPAG, ENIIGINDI, INDIIRTPT, and NDIIRTPTI) for further evaluation to determine their potential efficacy, safety, and suitability for vaccine design based on strong binding potential. The potential to cover a large portion of the world's population with these vaccines is substantial—88.5 % for one type and 99.99 % for another. In exploring the molecular docking analyses, we examined a library of compounds from the ZINC database. Among them, we identified twelve compounds with the lowest binding energy. Critical residues in the MAPK1 protein, such as VAL48, LYS63, CYS175, ASP176, LYS160, ALA61, LEU165, TYR45, SER162, ARG33, PRO365, PHE363, ILE40, ASN163, and GLU42, are pivotal for interactions with these compounds. Our result suggests that these compounds could influence the protein's behaviour. Moreover, our docking analyses revealed that the predicted peptides have a strong affinity for the MAPK1 protein. These peptides form stable complexes, indicating their potential as potent inhibitors. This study contributes to the identification of new drug compounds and the screening of their desired properties. These compounds could potentially help reduce the excessive activity of MAPK1, which is linked to Alzheimer's disease.
Graphical abstract
Related collections
Most cited references64

- Record: found
- Abstract: found
- Article: found
VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines
- Record: found
- Abstract: found
- Article: not found
Potential for primary prevention of Alzheimer's disease: an analysis of population-based data.

- Record: found
- Abstract: found
- Article: found