- Record: found
- Abstract: found
- Article: found
Novel molecular approach to define pest species status and tritrophic interactions from historical Bemisia specimens
Read this article at
Abstract
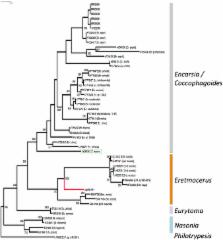
Museum specimens represent valuable genomic resources for understanding host-endosymbiont/parasitoid evolutionary relationships, resolving species complexes and nomenclatural problems. However, museum collections suffer DNA degradation, making them challenging for molecular-based studies. Here, the mitogenomes of a single 1912 Sri Lankan Bemisia emiliae cotype puparium, and of a 1942 Japanese Bemisia puparium are characterised using a Next-Generation Sequencing approach. Whiteflies are small sap-sucking insects including B. tabaci pest species complex. Bemisia emiliae’s draft mitogenome showed a high degree of homology with published B. tabaci mitogenomes, and exhibited 98–100% partial mitochondrial DNA Cytochrome Oxidase I (mtCOI) gene identity with the B. tabaci species known as Asia II-7. The partial mtCOI gene of the Japanese specimen shared 99% sequence identity with the Bemisia ‘JpL’ genetic group. Metagenomic analysis identified bacterial sequences in both Bemisia specimens, while hymenopteran sequences were also identified in the Japanese Bemisia puparium, including complete mtCOI and rRNA genes, and various partial mtDNA genes. At 88–90% mtCOI sequence identity to Aphelinidae wasps, we concluded that the 1942 Bemisia nymph was parasitized by an Eretmocerus parasitoid wasp. Our approach enables the characterisation of genomes and associated metagenomic communities of museum specimens using 1.5 ng gDNA, and to infer historical tritrophic relationships in Bemisia whiteflies.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Asymmetric mating interactions drive widespread invasion and displacement in a whitefly.
- Record: found
- Abstract: found
- Article: not found
Symbiont-mediated protection in insect hosts.
- Record: found
- Abstract: found
- Article: not found