- Record: found
- Abstract: found
- Article: found
Analysis of the 4q35 chromatin organization reveals distinct long-range interactions in patients affected with Facio-Scapulo-Humeral Dystrophy
Read this article at
Abstract
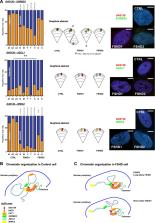
Facio-Scapulo Humeral dystrophy (FSHD) is the third most common myopathy, affecting 1 amongst 10,000 individuals (FSHD1, OMIM #158900). This autosomal dominant pathology is associated in 95% of cases with genetic and epigenetic alterations in the subtelomeric region at the extremity of the long arm of chromosome 4 (q arm). A large proportion of the remaining 5% of cases carry a mutation in the SMCHD1 gene (FSHD2, OMIM #158901). Here, we explored the 3D organization of the 4q35 locus by three-dimensions DNA in situ fluorescent hybridization (3D-FISH) in primary fibroblasts isolated from patients and healthy donors. We found that D4Z4 contractions and/or SMCHD1 mutations impact the spatial organization of the 4q35 region and trigger changes in the expression of different genes. Changes in gene expression were corroborated in muscle biopsies suggesting that the modified chromatin landscape impelled a modulation in the level of expression of a number of genes across the 4q35 locus in FSHD. Using induced pluripotent stem cells (hIPSC), we further examined whether chromatin organization is inherited after reprogramming or acquired during differentiation and showed that folding of the 4q35 region is modified upon differentiation. These results together with previous findings highlight the role of the D4Z4 macrosatellite repeat in the topological organization of chromatin and further indicate that the D4Z4-dependent 3D structure induces transcriptional changes of 4q35 genes expression.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Stability and flexibility of epigenetic gene regulation in mammalian development.
- Record: found
- Abstract: found
- Article: not found
Architectural protein subclasses shape 3D organization of genomes during lineage commitment.
- Record: found
- Abstract: found
- Article: not found