- Record: found
- Abstract: found
- Article: found
Alteration of the microRNA network during the progression of Alzheimer's disease
Read this article at
Abstract
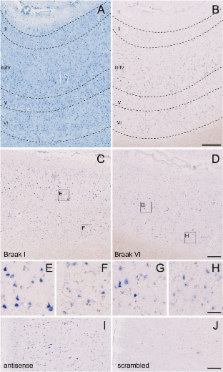
An overview of miRNAs altered in Alzheimer's disease (AD) was established by profiling the hippocampus of a cohort of 41 late-onset AD (LOAD) patients and 23 controls, showing deregulation of 35 miRNAs. Profiling of miRNAs in the prefrontal cortex of a second independent cohort of 49 patients grouped by Braak stages revealed 41 deregulated miRNAs. We focused on miR-132-3p which is strongly altered in both brain areas. Downregulation of this miRNA occurs already at Braak stages III and IV, before loss of neuron-specific miRNAs. Next-generation sequencing confirmed a strong decrease of miR-132-3p and of three family-related miRNAs encoded by the same miRNA cluster on chromosome 17. Deregulation of miR-132-3p in AD brain appears to occur mainly in neurons displaying Tau hyper-phosphorylation. We provide evidence that miR-132-3p may contribute to disease progression through aberrant regulation of mRNA targets in the Tau network. The transcription factor (TF) FOXO1a appears to be a key target of miR-132-3p in this pathway.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis.
- Record: found
- Abstract: found
- Article: not found