- Record: found
- Abstract: found
- Article: found
The methionine salvage pathway-involving ADI1 inhibits hepatoma growth by epigenetically altering genes expression via elevating S-adenosylmethionine

Read this article at
Abstract
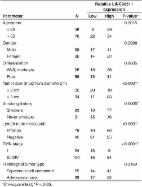
The 5′-methylthioadenosine (MTA) cycle-participating human acireductone dioxygenase 1 (ADI1) has been implicated as a tumor suppressor in prostate cancer, yet its role remains unclear in hepatocellular carcinoma (HCC). Here, we demonstrated a significant reduction of ADI1, either in protein or mRNA level, in HCC tissues. Additionally, higher ADI1 levels were associated with favorable postoperative recurrence-free survival in HCC patients. By altering ADI1 expression in HCC cells, a negative correlation between ADI1 and cell proliferation was observed. Cell-based and xenograft experiments were performed by using cells overexpressing ADI1 mutants carrying mutations at the metal-binding sites (E94A and H133A, respectively), which selectively disrupted differential catalytic steps, resulting in staying or leaving the MTA cycle. The results showed that the growth suppression effect was mediated by accelerating the MTA cycle. A cDNA microarray analysis followed by verification experiments identified that caveolin-1 (CAV1), a growth-promoting protein in HCC, was markedly decreased upon ADI1 overexpression. Suppression of CAV1 expression was mediated by an increase of S-adenosylmethionine (SAMe) level. The methylation status of CAV1 promoter was significantly altered upon ADI1 overexpression. Finally, a genome-wide methylation analysis revealed that ADI1 overexpression altered promoter methylation profiles in a set of cancer-related genes, including CAV1 and genes encoding antisense non-coding RNAs, long non-coding RNAs, and microRNAs, resulting in significant changes of their expression levels. In conclusion, ADI1 expression promoted MTA cycle to increase SAMe levels, which altered genome-wide promoter methylation profiles, resulting in altered gene expression and HCC growth suppression.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: found
Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non–small-cell Lung Cancer by Binding to EZH2 and Suppressing p57
- Record: found
- Abstract: found
- Article: not found
A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation.
- Record: found
- Abstract: found
- Article: not found