- Record: found
- Abstract: found
- Article: found
An evolved pyrrolysyl-tRNA synthetase with polysubstrate specificity expands the toolbox for engineering enzymes with incorporation of noncanonical amino acids
Read this article at
Abstract
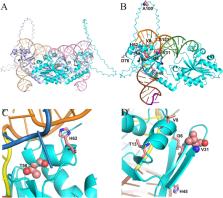
Aminoacyl-tRNA synthetase (aaRS) is a core component for genetic code expansion (GCE), a powerful technique that enables the incorporation of noncanonical amino acids (ncAAs) into a protein. The aaRS with polyspecificity can be exploited in incorporating additional ncAAs into a protein without the evolution of new, orthogonal aaRS/tRNA pair, which hence provides a useful tool for probing the enzyme mechanism or expanding protein function. A variant (N346A/C348A) of pyrrolysyl-tRNA synthetase from Methanosarcina mazei ( MmPylRS) exhibited a wide substrate scope of accepting over 40 phenylalanine derivatives. However, for most of the substrates, the incorporation efficiency was low. Here, a MbPylRS (N311A/C313A) variant was constructed that showed higher ncAA incorporation efficiency than its homologous MmPylRS (N346A/C348A). Next, N-terminal of MbPylRS (N311A/C313A) was engineered by a greedy combination of single variants identified previously, resulting in an IPE (N311A/C313A/V31I/T56P/A100E) variant with significantly improved activity against various ncAAs. Activity of IPE was then tested toward 43 novel ncAAs, and 16 of them were identified to be accepted by the variant. The variant hence could incorporate nearly 60 ncAAs in total into proteins. With the utility of this variant, eight various ncAAs were then incorporated into a lanthanide-dependent alcohol dehydrogenase PedH. Incorporation of phenyllactic acid improved the catalytic efficiency of PedH toward methanol by 1.8-fold, indicating the role of modifying protein main chain in enzyme engineering. Incorporation of O-tert-Butyl-L-tyrosine modified the enantioselectivity of PedH by influencing the interactions between substrate and protein. Enzymatic characterization and molecular dynamics simulations revealed the mechanism of ncAAs affecting PedH catalysis. This study provides a PylRS variant with high activity and substrate promiscuity, which increases the utility of GCE in enzyme mechanism illustration and engineering.
Related collections
Most cited references42
- Record: found
- Abstract: not found
- Article: not found
GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers
- Record: found
- Abstract: found
- Article: not found
CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data.
- Record: found
- Abstract: found
- Article: not found
