- Record: found
- Abstract: found
- Article: found
RecG Directs DNA Synthesis during Double-Strand Break Repair
Read this article at
Abstract
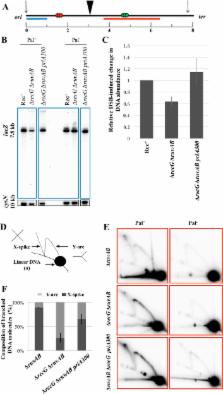
Homologous recombination provides a mechanism of DNA double-strand break repair (DSBR) that requires an intact, homologous template for DNA synthesis. When DNA synthesis associated with DSBR is convergent, the broken DNA strands are replaced and repair is accurate. However, if divergent DNA synthesis is established, over-replication of flanking DNA may occur with deleterious consequences. The RecG protein of Escherichia coli is a helicase and translocase that can re-model 3-way and 4-way DNA structures such as replication forks and Holliday junctions. However, the primary role of RecG in live cells has remained elusive. Here we show that, in the absence of RecG, attempted DSBR is accompanied by divergent DNA replication at the site of an induced chromosomal DNA double-strand break. Furthermore, DNA double-stand ends are generated in a recG mutant at sites known to block replication forks. These double-strand ends, also trigger DSBR and the divergent DNA replication characteristic of this mutant, which can explain over-replication of the terminus region of the chromosome. The loss of DNA associated with unwinding joint molecules previously observed in the absence of RuvAB and RecG, is suppressed by a helicase deficient PriA mutation ( priA300), arguing that the action of RecG ensures that PriA is bound correctly on D-loops to direct DNA replication rather than to unwind joint molecules. This has led us to put forward a revised model of homologous recombination in which the re-modelling of branched intermediates by RecG plays a fundamental role in directing DNA synthesis and thus maintaining genomic stability.
Author Summary
DNA double-strand breaks are accurately repaired by homologous recombination. This accuracy is ensured by copying the correct genetic information present on a second unbroken copy of the DNA, normally a sister chromosome that is generated during DNA replication. This implies that DNA synthesis occurring during recombination must be directed to replace lost or damaged base pairs but not to over-replicate undamaged chromosomal regions. Here, we investigate the genomic consequences of the absence of RecG during DNA repair following a site-specific double-strand break introduced in only one of two homologous E. coli chromosomes. Our observations suggest that RecG can re-model branched intermediates of recombination to direct the correct binding of PriA. This establishes converging replication forks that replace lost DNA at the site of DSBR and prevents over-replication of flanking DNA regions. This has led us to re-evaluate our understanding of the pathway of homologous recombination in E. coli and to propose a model in which RecG plays a critical role in remodelling branched intermediates at the interface of recombination and DNA replication.
Related collections
Most cited references59

- Record: found
- Abstract: found
- Article: found
The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets
- Record: found
- Abstract: found
- Article: not found
Replication fork stalling at natural impediments.
- Record: found
- Abstract: found
- Article: not found