- Record: found
- Abstract: found
- Article: not found
FGF4 retrogene on CFA12 is responsible for chondrodystrophy and intervertebral disc disease in dogs
Read this article at
Significance
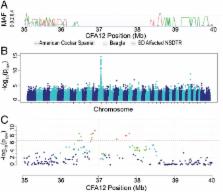
Chondrodystrophy, characterized by short limbs and intervertebral disc disease (IVDD), is a common phenotype in many of the most popular dog breeds, including the dachshund, beagle, and French bulldog. Here, we report the identification of a FGF4 retrogene insertion on chromosome 12, the second FGF4 retrogene reported in the dog, as responsible for chondrodystrophy and IVDD. Identification of the causative mutation for IVDD will impact an incredibly large proportion of the dog population and provides a model for IVDD in humans, as FGF-associated mutations are responsible for IVDD and short stature in human achondroplasia. This is a report of a second retrogene copy of the same parental gene, each causing complementary disease phenotypes in a mammalian species.
Abstract
Chondrodystrophy in dogs is defined by dysplastic, shortened long bones and premature degeneration and calcification of intervertebral discs. Independent genome-wide association analyses for skeletal dysplasia (short limbs) within a single breed ( P Bonferroni = 0.01) and intervertebral disc disease (IVDD) across breeds ( P Bonferroni = 4.0 × 10 −10) both identified a significant association to the same region on CFA12. Whole genome sequencing identified a highly expressed FGF4 retrogene within this shared region. The FGF4 retrogene segregated with limb length and had an odds ratio of 51.23 (95% CI = 46.69, 56.20) for IVDD. Long bone length in dogs is a unique example of multiple disease-causing retrocopies of the same parental gene in a mammalian species. FGF signaling abnormalities have been associated with skeletal dysplasia in humans, and our findings present opportunities for both selective elimination of a medically and financially devastating disease in dogs and further understanding of the ever-growing complexity of retrogene biology.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Active human retrotransposons: variation and disease.
- Record: found
- Abstract: found
- Article: found
Nosology and Classification of Genetic Skeletal Disorders: 2010 Revision
- Record: found
- Abstract: found
- Article: not found
