- Record: found
- Abstract: found
- Article: found
Genomic Insights into the Increased Occurrence of Campylobacteriosis Caused by Antimicrobial-Resistant Campylobacter coli
Read this article at
ABSTRACT
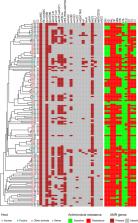
Campylobacter is the leading bacterial cause of diarrheal illnesses worldwide. Campylobacter jejuni and C. coli are the most common species accounting for campylobacteriosis. Although the proportion of campylobacteriosis caused by C. coli is increasing rapidly in China, the underlying mechanisms of this emergence remain unclear. In this study, we analyzed the whole-genome sequences and associated environments of 1,195 C. coli isolates with human, poultry, or porcine origins from 1980 to 2021. C. coli isolates of human origin were closely related to those from poultry, suggesting that poultry was the main source of C. coli infection in humans. Analysis of antimicrobial resistance determinants indicated that the prevalence of multidrug-resistant C. coli has increased dramatically since the 2010s, coinciding with the shift in abundance from C. jejuni to C. coli in Chinese poultry. Compared with C. jejuni, drug-resistant C. coli strains were better adapted and showed increased proliferation in the poultry production environment, where multiple antimicrobial agents were frequently used. This study provides an empirical basis for the molecular mechanisms that have enabled C. coli to become the dominant Campylobacter species in poultry; we also emphasize the importance of poultry products as sources of campylobacteriosis caused by C. coli in human patients.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: found
Identification of acquired antimicrobial resistance genes
- Record: found
- Abstract: found
- Article: not found