- Record: found
- Abstract: found
- Article: found
Loss-of-function variants affecting the STAGA complex component SUPT7L cause a developmental disorder with generalized lipodystrophy
Read this article at
Abstract
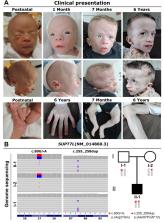
Generalized lipodystrophy is a feature of various hereditary disorders, often leading to a progeroid appearance. In the present study we identified a missense and a frameshift variant in a compound heterozygous state in SUPT7L in a boy with intrauterine growth retardation, generalized lipodystrophy, and additional progeroid features. SUPT7L encodes a component of the transcriptional coactivator complex STAGA. By transcriptome sequencing, we showed the predicted missense variant to cause aberrant splicing, leading to exon truncation and thereby to a complete absence of SUPT7L in dermal fibroblasts. In addition, we found altered expression of genes encoding DNA repair pathway components. This pathway was further investigated and an increased rate of DNA damage was detected in proband-derived fibroblasts and genome-edited HeLa cells. Finally, we performed transient overexpression of wildtype SUPT7L in both cellular systems, which normalizes the number of DNA damage events. Our findings suggest SUPT7L as a novel disease gene and underline the link between genome instability and progeroid phenotypes.
Related collections
Most cited references52

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found