- Record: found
- Abstract: found
- Article: found
FBXW7 in Cancer: What Has Been Unraveled Thus Far?
Read this article at
Abstract
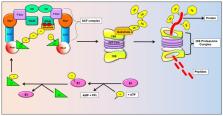
The FBXW7 (F-box with 7 tandem WD40) protein encoded by the gene FBXW7 is one of the crucial components of ubiquitin ligase called Skp1-Cullin1-F-box (SCF) complex that aids in the degradation of many oncoproteins via the ubiquitin-proteasome system (UPS) thus regulating cellular growth. FBXW7 is considered as a potent tumor suppressor as most of its target substrates can function as potential growth promoters, including c-Myc, Notch, cyclin E, c-JUN, and KLF5. Its regulators include p53, C/EBP-δ, Numb, microRNAs, Pin 1, Hes-5, BMI1, Ebp2. Mounting evidence has indicated the involvement of aberrant expression of FBXW7 for tumorigenesis. Moreover, numerous studies have also shown its role in cancer cell chemosensitization, thereby demonstrating the importance of FBXW7 in the development of curative cancer therapy. This comprehensive review emphasizes on the targets, functions, regulators and expression of FBXW7 in different cancers and its involvement in sensitizing cancer cells to chemotherapeutic drugs.
Related collections
Most cited references194
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
- Record: found
- Abstract: found
- Article: not found
FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation.
- Record: found
- Abstract: found
- Article: not found