- Record: found
- Abstract: found
- Article: not found
Transcriptional Silencing of Transposons by Piwi and Maelstrom and Its Impact on Chromatin State and Gene Expression

Read this article at
Summary
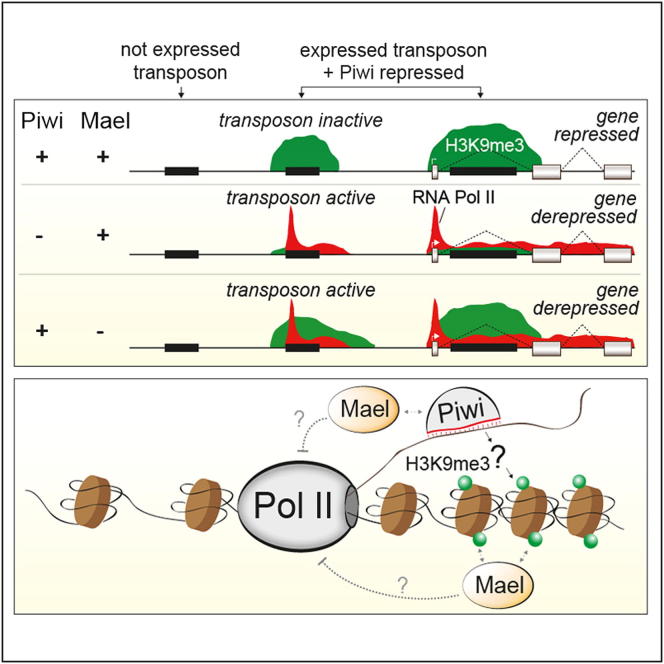
Eukaryotic genomes are colonized by transposons whose uncontrolled activity causes genomic instability. The piRNA pathway silences transposons in animal gonads, yet how this is achieved molecularly remains controversial. Here, we show that the HMG protein Maelstrom is essential for Piwi-mediated silencing in Drosophila. Genome-wide assays revealed highly correlated changes in RNA polymerase II recruitment, nascent RNA output, and steady-state RNA levels of transposons upon loss of Piwi or Maelstrom. Our data demonstrate piRNA-mediated trans-silencing of hundreds of transposon copies at the transcriptional level. We show that Piwi is required to establish heterochromatic H3K9me3 marks on transposons and their genomic surroundings. In contrast, loss of Maelstrom affects transposon H3K9me3 patterns only mildly yet leads to increased heterochromatin spreading, suggesting that Maelstrom acts downstream of or in parallel to H3K9me3. Our work illustrates the widespread influence of transposons and the piRNA pathway on chromatin patterns and gene expression.
Abstract
Highlights
► Piwi-RISC guides transcriptional silencing of transposons in trans ► Piwi-mediated silencing triggers H3K9me3 heterochromatin formation ► Maelstrom is required for transcriptional silencing, but not for H3K9 trimethylation ► Transposon silencing by the piRNA pathway broadly affects gene expression
Abstract
The piRNA-interacting protein Piwi and the HMG protein Maelstrom collaborate genome-wide to silence transcription of transposons in part through trimethylation of H3K9, which depends on Piwi. Silencing broadly affects the expression of genes neighboring the transposons, and Maelstrom restricts spreading of the repressive histone mark.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Mobile elements: drivers of genome evolution.
- Record: found
- Abstract: found
- Article: not found
Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary.
- Record: found
- Abstract: found
- Article: not found