- Record: found
- Abstract: found
- Article: found
Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways
Read this article at
Abstract
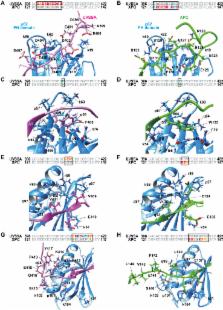
Nucleotide excision repair is initiated by two different damage recognition subpathways, global genome repair (GGR) and transcription-coupled repair (TCR). In GGR, XPC detects DNA lesions and recruits TFIIH via interaction with the pleckstrin homology (PH) domain of TFIIH subunit p62. In TCR, an elongating form of RNA Polymerase II detects a lesion on the transcribed strand and recruits TFIIH by an unknown mechanism. Here, we found that the TCR initiation factor UVSSA forms a stable complex with the PH domain of p62 via a short acidic string in the central region of UVSSA, and determined the complex structure by NMR. The acidic string of UVSSA binds strongly to the basic groove of the PH domain by inserting Phe408 and Val411 into two pockets, highly resembling the interaction mechanism of XPC with p62. Mutational binding analysis validated the structure and identified residues crucial for binding. TCR activity was markedly diminished in UVSSA-deficient cells expressing UVSSA mutated at Phe408 or Val411. Thus, a common TFIIH recruitment mechanism is shared by UVSSA in TCR and XPC in GGR.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Transcription-coupled DNA repair: two decades of progress and surprises.
- Record: found
- Abstract: found
- Article: not found
Protein backbone angle restraints from searching a database for chemical shift and sequence homology.
- Record: found
- Abstract: found
- Article: not found