- Record: found
- Abstract: found
- Article: found
Extensive alternative splicing transitions during postnatal skeletal muscle development are required for calcium handling functions
Read this article at
Abstract
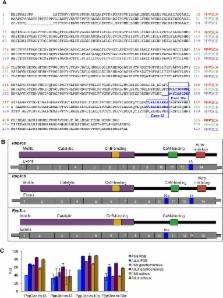
Postnatal development of skeletal muscle is a highly dynamic period of tissue remodeling. Here, we used RNA-seq to identify transcriptome changes from late embryonic to adult mouse muscle and demonstrate that alternative splicing developmental transitions impact muscle physiology. The first 2 weeks after birth are particularly dynamic for differential gene expression and alternative splicing transitions, and calcium-handling functions are significantly enriched among genes that undergo alternative splicing. We focused on the postnatal splicing transitions of the three calcineurin A genes, calcium-dependent phosphatases that regulate multiple aspects of muscle biology. Redirected splicing of calcineurin A to the fetal isoforms in adult muscle and in differentiated C2C12 slows the timing of muscle relaxation, promotes nuclear localization of calcineurin target Nfatc3, and/or affects expression of Nfatc transcription targets. The results demonstrate a previously unknown specificity of calcineurin isoforms as well as the broader impact of alternative splicing during muscle postnatal development.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
The evolutionary landscape of alternative splicing in vertebrate species.
- Record: found
- Abstract: found
- Article: not found
Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage.
- Record: found
- Abstract: found
- Article: not found