- Record: found
- Abstract: found
- Article: found
KS-CMI: A circRNA-miRNA interaction prediction method based on the signed graph neural network and denoising autoencoder
Read this article at
Summary
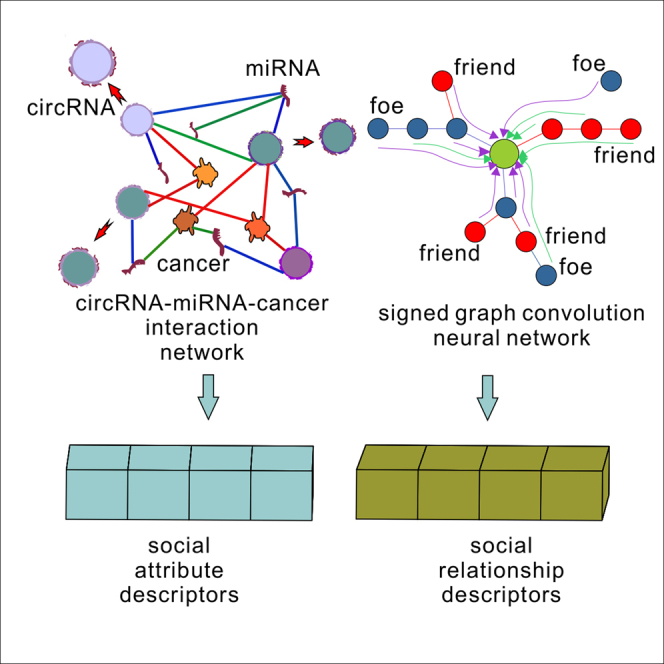
Circular RNA (circRNA) plays an important role in the diagnosis, treatment, and prognosis of human diseases. The discovery of potential circRNA-miRNA interactions (CMI) is of guiding significance for subsequent biological experiments. Limited by the small amount of experimentally supported data and high randomness, existing models are difficult to accomplish the CMI prediction task based on real cases. In this paper, we propose KS-CMI, a novel method for effectively accomplishing CMI prediction in real cases. KS-CMI enriches the ‘behavior relationships’ of molecules by constructing circRNA-miRNA-cancer (CMCI) networks and extracts the behavior relationship attribute of molecules based on balance theory. Next, the denoising autoencoder (DAE) is used to enhance the feature representation of molecules. Finally, the CatBoost classifier was used for prediction. KS-CMI achieved the most reliable prediction results in real cases and achieved competitive performance in all datasets in the CMI prediction.
Graphical abstract
Highlights
Abstract
Gene network; Neural networks
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
Detecting and characterizing circular RNAs.

- Record: found
- Abstract: found
- Article: not found
Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.