- Record: found
- Abstract: found
- Article: found
Reduced polymorphism of Plasmodium vivax early transcribed membrane protein (PvETRAMP) 11.2
Read this article at
Abstract
Background
ETRAMP11.2 (PVX_003565) is a well-characterized protein with antigenic potential. It is considered to be a serological marker for diagnostic tools, and it has been suggested as a potential vaccine candidate. Despite its immunological relevance, the polymorphism of the P. vivax ETRAMP11.2 gene ( pvetramp11.2) remains undefined. The genetic variability of an antigen may limit the effectiveness of its application as a serological surveillance tool and in vaccine development and, therefore, the aim of this study was to investigate the genetic diversity of pvetramp11.2 in parasite populations from Amazonian regions and worldwide. We also evaluated amino acid polymorphism on predicted B-cell epitopes. The low variability of the sequence encoding PvETRAMP11.2 protein suggests that it would be a suitable marker in prospective serodiagnostic assays for surveillance strategies or in vaccine design against P. vivax malaria.
Methods
The pvetramp11.2 of P. vivax isolates collected from Brazil ( n = 68) and Peru ( n = 36) were sequenced and analyzed to assess nucleotide polymorphisms, allele distributions, population differentiation, genetic diversity and signature of selection. In addition, sequences ( n = 104) of seven populations from different geographical regions were retrieved from the PlasmoDB database and included in the analysis to study the worldwide allele distribution. Potential linear B-cell epitopes and their polymorphisms were also explored.
Results
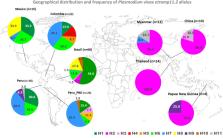
The multiple alignments of 208 pvetramp11.2 sequences revealed a low polymorphism and a marked geographical variation in allele diversity. Seven polymorphic sites and 11 alleles were identified. All of the alleles were detected in isolates from the Latin American region and five alleles were detected in isolates from the Southeast Asia/Papua New Guinea (SEA/PNG) region. Three alleles were shared by all Latin American populations (H1, H6 and H7). The H1 allele (reference allele from Salvador-1 strain), which was absent in the SEA/PNG populations, was the most represented allele in populations from Brazil (54%) and was also detected at high frequencies in populations from all other Latin America countries (range: 13.0% to 33.3%). The H2 allele was the major allele in SEA/PNG populations, but was poorly represented in Latin America populations (only in Brazil: 7.3%). Plasmodium vivax populations from Latin America showed a marked inter-population genetic differentiation (fixation index [Fst]) in contrast to SEA/PNG populations. Codon bias measures (effective number of codons [ENC] and Codon bias index [CBI]) indicated preferential use of synonymous codons, suggesting selective pressure at the translation level. Only three amino acid substitutions, located in the C-terminus, were detected. Linear B-cell epitope mapping predicted two epitopes in the Sal-1 PvETRAMP11.2 protein, one of which was fully conserved in all of the parasite populations analyzed.
Conclusions
We provide an overview of the allele distribution and genetic differentiation of ETRAMP11.2 antigen in P. vivax populations from different endemic areas of the world. The reduced polymorphism and the high degree of protein conservation supports the application of PvETRAMP11.2 protein as a reliable antigen for application in serological assays or vaccine design. Our findings provide useful information that can be used to inform future study designs.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets.
- Record: found
- Abstract: found
- Article: not found
DnaSP v5: a software for comprehensive analysis of DNA polymorphism data.
- Record: found
- Abstract: found
- Article: found
