- Record: found
- Abstract: found
- Article: found
Data on the DNA damaging and mutagenic potential of the BH3-mimetics ABT-263/Navitoclax and TW-37
Read this article at
Abstract
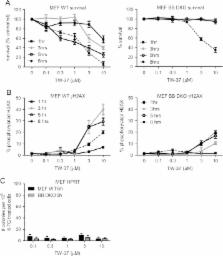
Unfortunately, the mutagenic activities of chemotherapy and radiotherapy can provoke development of therapy-induced malignancies in cancer survivors. Non-mutagenic anti-cancer therapies may be less likely to trigger subsequent malignant neoplasms. Here we present data regarding the DNA damaging and mutagenic potential of two drugs that antagonize proteins within the Bcl-2 family: ABT-263/Navitoclax and TW-37. Our data reveal that concentrations of these agents that stimulated Bax/Bak-dependent signaling provoked little DNA damage and failed to trigger mutations in surviving cells. The data supplied in this article is related to the research work entitled "Inhibition of Bcl-2 or IAP proteins does not provoke mutations in surviving cells" [1].
Related collections
Most cited references5
- Record: found
- Abstract: found
- Article: not found
Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays.

- Record: found
- Abstract: found
- Article: found
TRAIL treatment provokes mutations in surviving cells
- Record: found
- Abstract: found
- Article: not found