- Record: found
- Abstract: found
- Article: found
Local and global influences on protein turnover in neurons and glia
Read this article at
Abstract
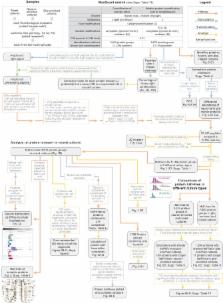
Regulation of protein turnover allows cells to react to their environment and maintain homeostasis. Proteins can show different turnover rates in different tissue, but little is known about protein turnover in different brain cell types. We used dynamic SILAC to determine half-lives of over 5100 proteins in rat primary hippocampal cultures as well as in neuron-enriched and glia-enriched cultures ranging from <1 to >20 days. In contrast to synaptic proteins, membrane proteins were relatively shorter-lived and mitochondrial proteins were longer-lived compared to the population. Half-lives also correlate with protein functions and the dynamics of the complexes they are incorporated in. Proteins in glia possessed shorter half-lives than the same proteins in neurons. The presence of glia sped up or slowed down the turnover of neuronal proteins. Our results demonstrate that both the cell-type of origin as well as the nature of the extracellular environment have potent influences on protein turnover.
Related collections
Most cited references35

- Record: found
- Abstract: found
- Article: found
The Proteomics Identifications (PRIDE) database and associated tools: status in 2013

- Record: found
- Abstract: found
- Article: found
CORUM: the comprehensive resource of mammalian protein complexes—2009
- Record: found
- Abstract: found
- Article: not found