- Record: found
- Abstract: found
- Article: not found
Distinct stem/progenitor cells proliferate to regenerate the trachea, intrapulmonary airways and alveoli in COVID-19 patients
letter
Yinshan Fang
1 ,
Helu Liu
1 ,
Huachao Huang
1 ,
Haiyan Li
2 ,
Anjali Saqi
3 ,
Li Qiang
3 ,
Jianwen Que
1
,
30 June 2020
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Dear Editor,
The global pandemic COVID-19 caused by SARS-CoV-2 virus has infected over 6.5 million
individuals and claimed over 350,000 lives worldwide within six months. The respiratory
epithelial cells covering the airways and alveoli are the major targets of the virus.
Moreover, damage to the epithelium can be exacerbated by mechanical ventilation. It
is expected that many of the infected individuals that survived the acute phase will
develop pulmonary diseases (e.g. fibrosis) if the epithelium fails to regenerate properly.
Multiple stem/progenitor cells have been implicated in the regeneration of the respiratory
epithelium. The human trachea and intrapulmonary airways are lined by three major
cell types, basal, club and ciliated cells, and the alveolar epithelium includes type
1 (AT1) and type 2 (AT2) cells. The trachea basal cells have been shown to serve as
progenitor cells to self-renew and differentiate into other cell types including club,
ciliated cells and minor cell populations (e.g., tuft cells).
1,2
Club cells can also de-differentiate into basal cells to regenerate the tracheal epithelium
in a mouse model where basal cells are ablated prior to injury.
3
In the intrapulmonary airways, basal cells have been postulated to serve as progenitor
cells for epithelial regeneration in humans (reviewed by
4
). In mice, however, club cells are responsible for repopulating the intrapulmonary
airway epithelium upon injury due to their lack of basal cells in the intrapulmonary
airways.
5
The cell of origin for regenerating the alveolar epithelium remains controversial.
Multiple cell types have been implicated in the regeneration of the alveolar epithelium
depending on injury models. These cells include AT1 and AT2 cells, bronchial-alveolar
ductal cells (BASCs), distal airway stem cells (DASCs), lineage negative epithelial
precursor (LNEP) cells, bronchial epithelial stem cells (BESCs), and different AT2
subpopulation cells (reviewed by
6
). Although histology analysis has been performed, it remains unknown which stem/progenitor
cell(s) proliferate in response to viral challenges in COVID-19 patients.
Cellular entry of SARS-CoV-2 depends on the extracellular receptor Angiotensin Converting
Enzyme 2 (ACE2) and the serine protease transmembrane serine protease 2 (TMPRSS2).
ACE2 and TMPRSS2 are expressed in both nasal and bronchial epithelium as detected
by immunohistochemistry.
7
Single cell RNA-sequencing analysis confirmed that ACE2 is enriched in the human airway
epithelium including club, ciliated and goblet cells, AT1 and AT2 cells.
8,9
ACE2 and TMPRSS2 are also co-expressed in a subpopulation of ciliated cells and AT2
cells.
8
Consistently, histology characterization revealed that SARS-CoV-2 infection induces
severe damages in the intrapulmonary airways and alveoli.
10
However, detailed characterization of different respiratory cell types remains lacking
for COVID-19 patients. In this report we showed that ciliated, club, AT1 and AT2 cells
are the major cell types damaged by SARS-CoV-2 infection. More importantly, we demonstrated
that distinct proliferating cells are present in the trachea/large airways, small
airways and alveoli following SARS-CoV-2 infection.
We examined tracheas and lungs from five deceased patients with postmortem intervals
(PMIs) as low as 2.5 hours. The epithelium was severely damaged in some parts of the
trachea (Supplementary information, Fig. S1a, b). Ciliated and club cells were shed
into the lumen, and the underlying basal cells were exposed (Fig. 1a, b and data not
shown). Although KRT5 remained to be expressed in these exposed basal cells, the nuclei
rounded up in contrast to the neighboring basal cells where the overlaying club and
ciliated were intact (Supplementary information, Fig. S1c). Intriguingly, although
basal cells and other epithelial cells rarely proliferate at homeostasis in the adults,
1
extensive basal cell proliferation was observed in the trachea, especially in the
area where club and ciliated cells were damaged (Fig. 1b). The proliferating cells
were limited to the immediate parabasal layer in the area where the epithelial integrity
was relatively well maintained (Fig. 1a). However, in the severely damaged area, proliferating
cells were occasionally observed in the basal layer which is lined by basal cells
with round nuclei (Fig. 1b). Together these findings demonstrate that in the trachea
basal cells proliferate, likely serving as progenitor cells to regenerate the damaged
epithelium following SARS-CoV-2 infection.
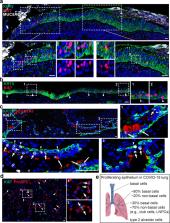
Fig. 1
Distinct proliferating cells in the trachea, intrapulmonary airways and alveoli of
COVID-19 patients.
a Proliferating basal cells are enriched in the area where severe damages occur. Note
Ki67+ cells are limited to the immediate parabasal layer. b Proliferating basal cells
are present in the basal (arrowheads) and immediate parabasal layers where the overlaying
club and ciliated are depleted. c Proliferating cells in the intrapulmonary airways
include basal cells (arrowheads), club cells (long arrows) and KRT5− SCGB1A1− population
(arrows). d Extensive proliferating alveolar type 2 cells (arrows). e Schematics of
proliferating epithelial cells in COVID-19 trachea and lung. Note the predominant
proliferating cells in the small airways are Lineage Negative Proliferating Cells
(LNPCs). In this study tracheas and lungs from five COVID-19 patients were examined.
Scale bar, 50 µm.
We next examined the COVID-19 lungs. The intrapulmonary airways and alveoli were severely
damaged with epithelial denudation and extensive intra-alveolar fibrinous exudates
(Supplementary information, Fig. S2a). A significant amount of ciliated and club cells
were sloughed and detached from basal cells in some areas (Supplementary information,
Fig. S2b). In the large airways (diameter > 0.5 mm), 82% of Ki67+ cells co-express
KRT5 (Supplementary information, Fig. S3a), suggesting that basal cell remains to
be the major proliferative cell population in response to viral challenge. By contrast,
in the small airways (diameter < 0.5 mm) although 29.08% ± 2.93% of basal cells showed
Ki67 expression, the majority of proliferating epithelial cells were KRT5− (4.10%
± 1.53% SCGB1A1+ KRT5− and 66.82% ± 3.31% SCGB1A1− KRT5−). Goblet cells (MUC5AC+)
and ciliated cells (FOXJ1+) were not proliferative (Supplementary information, Fig. S3a
and data not shown, n = 5). Although a previous study showed that approximately 1%
to 2% of neuroendocrine cells are proliferative,
11
we did not observe any Ki67+ neuroendocrine cells (Synaptophysin, SYP+) (Supplementary
information, Fig. S3b, 0/16 cells). While extensive CD45+ cells were present in the
mesenchyme, the inflammatory cells did not seem to infiltrate into the epithelial
layer (Supplementary information, Fig. S3c). We did not observe proliferating AT1
cells in the alveoli (Supplementary information, Fig. S4). 3.99% ± 1.66% of AT2 cells
exhibited Ki67 staining in the parenchyma where the alveoli were relatively well preserved.
However, in severely damaged area 15.69% ± 1.87% of AT2 cells were proliferative (Fig. 1d;
Supplementary information, Fig. S4), suggesting that AT2 cells are mobilized to regenerate
the alveoli.
Taken together, we demonstrate that distinct cell populations proliferate in different
regions of the respiratory system following SARS-CoV-2 infection (Fig. 1e). In the
trachea and larger airways basal cells (KRT5+) proliferate extensively. This is consistent
with a previous report showing that approximately 84% proliferating cells express
KRT5 in the normal large airways (>2 mm).
12
Interestingly, although approximately 30% proliferating cells exhibit KRT5 expression
in the small airways of COVID-19 lungs, the majority of proliferating cells do not
express KRT5. Among them the predominant cell population does not express lineage
markers including SCGB1A1, FOXJ1, acetylated tubulin, MUC5AC and SYP as evidenced
by immunostaining. Whether these Lineage Negative Proliferating Cells (LNPCs) are
similar to the LNEPs identified in mouse models
13
remains to be determined. It is also unknown whether these cells are derived from
the underlying basal cells or neighboring club cells. The alveoli AT2 cells proliferate
extensively in response to virus-induced damage. Mouse studies have shown that basal
cells and AT2 cells consist of multiple subpopulations (reviewed by
6
). It will be interesting to further determine which subpopulations are activated
and participate in lung regeneration in COVID-19 patients. Understanding the molecular
mechanisms that promote the expansion of distinct progenitors present in the proximal/distal
airways and alveoli is critical for rebuilding a functional respirator system.
Supplementary information
Supplementary, Figures and Methods
Related collections
Most cited references4
- Record: found
- Abstract: found
- Article: not found
Basal cells as stem cells of the mouse trachea and human airway epithelium.
Jason R. Rock, Mark Onaitis, Emma L. Rawlins … (2009)
- Record: found
- Abstract: found
- Article: not found
Number and proliferation of basal and parabasal cells in normal human airway epithelium.
F Thunnissen, Susanne J. Boers, Ton A Ambergen (1998)
- Record: found
- Abstract: found
- Article: not found
Airway basal cells. The "smoking gun" of chronic obstructive pulmonary disease.
Ronald G. Crystal (2014)
