- Record: found
- Abstract: found
- Article: found
DrugComb update: a more comprehensive drug sensitivity data repository and analysis portal

Read this article at
Abstract
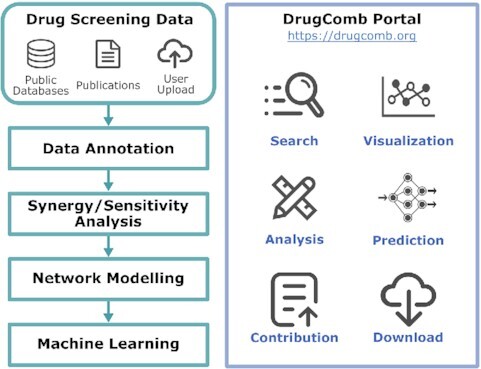
Combinatorial therapies that target multiple pathways have shown great promises for treating complex diseases. DrugComb ( https://drugcomb.org/) is a web-based portal for the deposition and analysis of drug combination screening datasets. Since its first release, DrugComb has received continuous updates on the coverage of data resources, as well as on the functionality of the web server to improve the analysis, visualization and interpretation of drug combination screens. Here, we report significant updates of DrugComb, including: (i) manual curation and harmonization of more comprehensive drug combination and monotherapy screening data, not only for cancers but also for other diseases such as malaria and COVID-19; (ii) enhanced algorithms for assessing the sensitivity and synergy of drug combinations; (iii) network modelling tools to visualize the mechanisms of action of drugs or drug combinations for a given cancer sample and (iv) state-of-the-art machine learning models to predict drug combination sensitivity and synergy. These improvements have been provided with more user-friendly graphical interface and faster database infrastructure, which make DrugComb the most comprehensive web-based resources for the study of drug sensitivities for multiple diseases.
Graphical Abstract
Related collections
Most cited references52

- Record: found
- Abstract: found
- Article: found
DrugBank 5.0: a major update to the DrugBank database for 2018

- Record: found
- Abstract: found
- Article: found