- Record: found
- Abstract: found
- Article: found
Functional Analysis of the Replication Fork Proteome Identifies BET Proteins as PCNA Regulators
Read this article at
SUMMARY
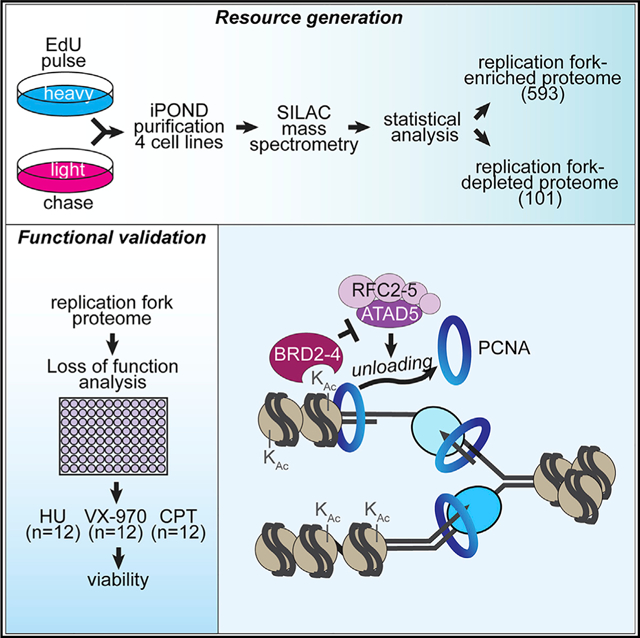
Identifying proteins that function at replication forks is essential to understanding DNA replication, chromatin assembly, and replication-coupled DNA repair mechanisms. Combining quantitative mass spectrometry in multiple cell types with stringent statistical cutoffs, we generated a high-confidence catalog of 593 proteins that are enriched at replication forks and nascent chromatin. Loss-of-function genetic analyses indicate that 85% yield phenotypes that are consistent with activities in DNA and chromatin replication or already have described functions in these processes. We illustrate the value of this resource by identifying activities of the BET family proteins BRD2, BRD3, and BRD4 in controlling DNA replication. These proteins use their extra-terminal domains to bind and inhibit the ATAD5 complex and thereby control the amount of PCNA on chromatin.
Graphical Abstract
In Brief
Wessel et al. use iPOND purifications and loss-of-function analyses to identify and characterize a replication fork and nascent chromatin proteome of 593 proteins. They demonstrate the resource’s utility by showing that the BET proteins BRD2, BRD3, and BRD4 inhibit ATAD5-dependent PCNA unloading from chromatin.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Regulated Eukaryotic DNA Replication Origin Firing with Purified Proteins
- Record: found
- Abstract: found
- Article: not found