- Record: found
- Abstract: found
- Article: found
Exploring Dynamic Changes in HIV-1 Molecular Transmission Networks and Key Influencing Factors: Cross-Sectional Study
Abstract
Background
The HIV-1 molecular network is an innovative tool, using gene sequences to understand transmission attributes and complementing social and sexual network studies. While previous research focused on static network characteristics, recent studies’ emphasis on dynamic features enhances our understanding of real-time changes, offering insights for targeted interventions and efficient allocation of public health resources.
Objective
This study aims to identify the dynamic changes occurring in HIV-1 molecular transmission networks and analyze the primary influencing factors driving the dynamics of HIV-1 molecular networks.
Methods
We analyzed and compared the dynamic changes in the molecular network over a specific time period between the baseline and observed end point. The primary factors influencing the dynamic changes in the HIV-1 molecular network were identified through univariate analysis and multivariate analysis.
Results
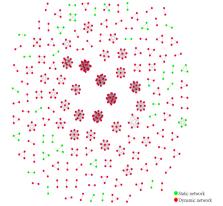
A total of 955 HIV-1 polymerase fragments were successfully amplified from 1013 specimens; CRF01_AE and CRF07_BC were the predominant subtypes, accounting for 40.8% (n=390) and 33.6% (n=321) of the specimens, respectively. Through the analysis and comparison of the basic and terminal molecular networks, it was discovered that 144 sequences constituted static molecular networks, and 487 sequences contributed to the formation of dynamic molecular networks. The findings of the multivariate analysis indicated that the factors occupation as a student, floating population, Han ethnicity, engagement in occasional or multiple sexual partnerships, participation in anal sex, and being single were independent risk factors for the dynamic changes observed in the HIV-1 molecular network, and the odds ratio (OR; 95% CIs) values were 2.63 (1.54-4.47), 1.83 (1.17-2.84), 2.91 (1.09-7.79), 1.75 (1.06-2.90), 4.12 (2.48-6.87), 5.58 (2.43-12.80), and 2.10 (1.25-3.54), respectively. Heterosexuality and homosexuality seem to exhibit protective effects when compared to bisexuality, with OR values of 0.12 (95% CI 0.05-0.32) and 0.26 (95% CI 0.11-0.64), respectively. Additionally, the National Eight-Item score and sex education experience were also identified as protective factors against dynamic changes in the HIV-1 molecular network, with OR values of 0.12 (95% CI 0.05-0.32) and 0.26 (95% CI 0.11-0.64), respectively.
Conclusions
The HIV-1 molecular network analysis showed 144 sequences in static networks and 487 in dynamic networks. Multivariate analysis revealed that occupation as a student, floating population, Han ethnicity, and risky sexual behavior were independent risk factors for dynamic changes, while heterosexuality and homosexuality were protective compared to bisexuality. A higher National Eight-Item score and sex education experience were also protective factors. The identification of HIV dynamic molecular networks has provided valuable insights into the characteristics of individuals undergoing dynamic alterations. These findings contribute to a better understanding of HIV-1 transmission dynamics and could inform targeted prevention strategies.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.

- Record: found
- Abstract: found
- Article: found
Automated analysis of phylogenetic clusters

- Record: found
- Abstract: found
- Article: found
Transmission Network Parameters Estimated From HIV Sequences for a Nationwide Epidemic
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.
