- Record: found
- Abstract: found
- Article: found
A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda
Read this article at
Abstract
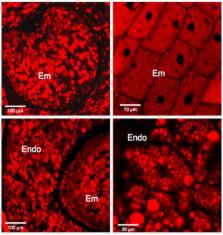
Desiccation tolerance allows plant seeds to remain viable in a dry state for years and even centuries. To reveal potential evolutionary processes of this trait, we have conducted a shotgun proteomic analysis of isolated embryo and endosperm from mature seeds of Amborella trichopoda, an understory shrub endemic to New Caledonia that is considered to be the basal extant angiosperm. The present analysis led to the characterization of 415 and 69 proteins from the isolated embryo and endosperm tissues, respectively. The role of these proteins is discussed in terms of protein evolution and physiological properties of the rudimentary, underdeveloped, Amborella embryos, notably considering that the acquisition of desiccation tolerance corresponds to the final developmental stage of mature seeds possessing large embryos.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
Seed germination and vigor.

- Record: found
- Abstract: found
- Article: found
The heat‐shock protein/chaperone network and multiple stress resistance
- Record: found
- Abstract: found
- Article: not found