- Record: found
- Abstract: found
- Article: found
Aptamers targeting amyloidogenic proteins and their emerging role in neurodegenerative diseases
Read this article at
Abstract
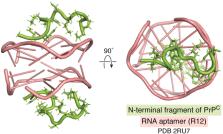
Aptamers are oligonucleotides selected from large pools of random sequences based on their affinity for bioactive molecules and are used in similar ways to antibodies. Aptamers provide several advantages over antibodies, including their small size, facile, large-scale chemical synthesis, high stability, and low immunogenicity. Amyloidogenic proteins, whose aggregation is relevant to neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and prion diseases, are among the most challenging targets for aptamer development due to their conformational instability and heterogeneity, the same characteristics that make drug development against amyloidogenic proteins difficult. Recently, chemical tethering of aptagens (equivalent to antigens) and advances in high-throughput sequencing-based analysis have been used to overcome some of these challenges. In addition, internalization technologies using fusion to cellular receptors and extracellular vesicles have facilitated central nervous system (CNS) aptamer delivery. In view of the development of these techniques and resources, here we review antiamyloid aptamers, highlighting preclinical application to CNS therapy.
Related collections
Most cited references162
- Record: found
- Abstract: found
- Article: not found
Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine
- Record: found
- Abstract: found
- Article: not found
Shedding light on the cell biology of extracellular vesicles
- Record: found
- Abstract: found
- Article: not found