- Record: found
- Abstract: found
- Article: found
Protein and lipid mass concentration measurement in tissues by stimulated Raman scattering microscopy
Read this article at
Significance
We report a quantitative Raman microscopy method that measures the concentration of protein and lipid in cells at high spatial resolution in living and in fixed samples of tissues, allowing quantitative studies of cell size and organelle regulation both in cell culture and in tissue slices; it can be applied to problems of cell size control, intracellular crowding, and lipid metabolism in the context of cell growth, cell differentiation, cell senescence, and pathology.
Abstract
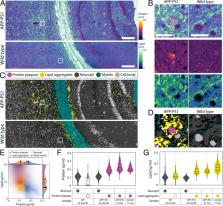
Cell mass and chemical composition are important aggregate cellular properties that are especially relevant to physiological processes, such as growth control and tissue homeostasis. Despite their importance, it has been difficult to measure these features quantitatively at the individual cell level in intact tissue. Here, we introduce normalized Raman imaging (NoRI), a stimulated Raman scattering (SRS) microscopy method that provides the local concentrations of protein, lipid, and water from live or fixed tissue samples with high spatial resolution. Using NoRI, we demonstrate that protein, lipid, and water concentrations at the single cell are maintained in a tight range in cells under the same physiological conditions and are altered in different physiological states, such as cell cycle stages, attachment to substrates of different stiffness, or by entering senescence. In animal tissues, protein and lipid concentration varies with cell types, yet an unexpected cell-to-cell heterogeneity was found in cerebellar Purkinje cells. The protein and lipid concentration profile provides means to quantitatively compare disease-related pathology, as demonstrated using models of Alzheimer’s disease. This demonstration shows that NoRI is a broadly applicable technique for probing the biological regulation of protein mass, lipid mass, and water mass for studies of cellular and tissue growth, homeostasis, and disease.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification.
- Record: found
- Abstract: found
- Article: not found