- Record: found
- Abstract: found
- Article: found
A haplotype-resolved reference genome of a long-distance migratory bat, Pipistrellus nathusii (Keyserling & Blasius, 1839)
Read this article at
Abstract
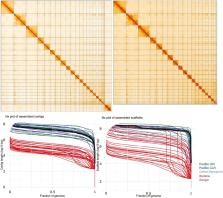
We present a complete, chromosome-scale reference genome for the long-distance migratory bat Pipistrellus nathusii. The genome encompasses both haplotypic sets of autosomes and the separation of both sex chromosomes by utilizing highly accurate long-reads and preserving long-range phasing information through the use of three-dimensional chromatin conformation capture sequencing (Hi-C). This genome, accompanied by a comprehensive protein-coding sequence annotation, provides a valuable genomic resource for future investigations into the genomic bases of long-distance migratory flight in bats as well as uncovering the genetic architecture, population structure and evolutionary history of Pipistrellus nathusii. The reference-quality genome presented here gives a fundamental resource to further our understanding of bat genetics and evolution, adding to the growing number of high-quality genetic resources in this field. Here, we demonstrate its use in the phylogenetic reconstruction of the order Chiroptera, and in particular, we present the resources to allow detailed investigations into the genetic drivers and adaptations related to long-distance migration.
Related collections
Most cited references53

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads
- Record: found
- Abstract: found
- Article: not found