- Record: found
- Abstract: found
- Article: found
Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy
Read this article at
Abstract
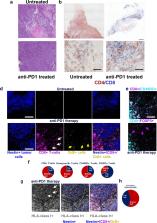
Anti-PD-1 therapy is used as a front-line treatment for many cancers, but mechanistic insight into this therapy resistance is still lacking. Here we generate a humanized (Hu)-mouse melanoma model by injecting fetal liver-derived CD34 + cells and implanting autologous thymus in immune-deficient NOD- scid IL2Rγ null (NSG) mice. Reconstituted Hu-mice are challenged with HLA-matched melanomas and treated with anti-PD-1, which results in restricted tumor growth but not complete regression. Tumor RNA-seq, multiplexed imaging and immunohistology staining show high expression of chemokines, as well as recruitment of FOXP3 + Treg and mast cells, in selective tumor regions. Reduced HLA-class I expression and CD8 +/Granz B + T cells homeostasis are observed in tumor regions where FOXP3 + Treg and mast cells co-localize, with such features associated with resistance to anti-PD-1 treatment. Combining anti-PD-1 with sunitinib or imatinib results in the depletion of mast cells and complete regression of tumors. Our results thus implicate mast cell depletion for improving the efficacy of anti-PD-1 therapy.
Abstract
Immune checkpoint therapies (ICT) are promising for treating various cancers, but response rates vary. Here the authors show, in mouse models, that tumor-infiltrating mast cells colocalize with regulatory T cells, coincide with local reduction of MHC-I and CD8 T cells, and is associated with resistance to ICT, which can be reversed by c-kit inhibitor treatment.
Related collections
Most cited references46

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy.
- Record: found
- Abstract: found
- Article: not found