- Record: found
- Abstract: found
- Article: found
Identification of methylated differentially expressed hub genes in systemic lupus erythematosus patients by WGCNA-based epigenome-wide and transcriptome-wide analysis
brief-report
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease, characterized by the
production of autoantibodies and the involvement of multi-systems. In order to explore
its molecular mechanism, we, using bioinformatics analysis and wet lab experiments,
identified two key genes in SLE patients.
Briefly, we analyzed differentially expressed genes (DEGs) and differentially methylated
genes (DMGs) from the GEO database (GSE50772 and GSE59250). The key genes were screened
by constructing a weighted gene co-expression network analysis (WGCNA) network. We
further performed immune infiltration analysis, functional enrichment analysis, and
motif transcriptional regulation analysis. Wet lab verification, including quantitative
real-time polymerase chain reaction (qPCR) and western blotting, was performed to
further confirm the expression of the identified key genes in SLE patients. The overall
workflow is shown in Figure S1A and the detailed research methods are in Supplementary
File 1.
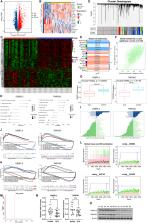
Based on the screening criteria (adjusted P < 0.05), 6,817 DEGs in GSE50772 were screened
(Fig. 1A). The top 10 upregulated or downregulated genes were clustered by visual
inspection of the heatmap (Fig. 1B). GO analysis indicated the enriched pathways of
these DEGs in each category (Fig. S1B). KEGG pathway enrichment showed that they were
mainly involved in herpes simplex virus 1 infection (Fig. S1B). Using the R package
“ChAMP”, 238 differential methylated probes (DMPs) were screened out from GSE59250
(Fig. 1C). By comparing the DMGs and the DEGs, we found four hypermethylated downregulated
genes and 35 hypomethylated upregulated genes (Fig. S1C). To determine which specific
genes are highly associated with SLE, we used the expression profile of GSE50772 to
construct a WGCNA network and explore the disease modules related to SLE. Samples
were first clustered to detect outliners and none was excluded (Fig. S1D). Soft threshold
power β = 12 (scale-free R
2 = 0.9) was selected (Fig. S1E). In total, 19,746 genes were classified into 18 distinct
modules based on the TOM matrix (Fig. 1D). Through further analysis between modules
and traits, we found that the light-green module had the highest correlation with
SLE phenotype (cor = −0.69, P = 1e−12; Fig. 1E). The correlation between this module
and the gene expression profiles was calculated. The gene significance score and module
membership in the light-green module was also highly correlated (cor = 0.6, P < 1e−200;
Fig. 1F). Therefore, this module was used for a further selection of key driver genes.
By intersecting the WGCNA light-green module genes with DEGs-DMGs (Fig. S1F), we obtained
two key genes OSBPL3 and TNRC6C (Fig. 1G). By analyzing the correlation between core
genes and immune infiltration in the dataset, we further explored their potential
molecular mechanisms in disease progression with regard to the immune component. We
quantified the immune infiltration level of each patient using the CIBERSORT algorithm
and found that both OSBPL3 and TNRC6C were significantly positively correlated with
CD4 memory T cells resting and negatively correlated with neutrophils and monocytes
(Fig. 1H). OSBPL3 was also positively correlated with γδT cells and NK cells resting,
while negatively correlated with activated dendritic cells (Fig. 1H). TNRC6C was positively
correlated with naïve CD4 T cells and naïve B cells, while negatively correlated with
memory B cells and plasma cells (Fig. 1H). The specific signal pathways involving
the core genes in the disease progression were investigated too. GSVA data demonstrated
that upregulated OSBPL3 was mainly enriched in G2M checkpoint and UV response DNA
pathways. Upregulated TNRC6C was mainly enriched in the G2M checkpoint and bile acid
metabolism pathways. Downregulated OSBPL3 and TNRC6C were both mainly enriched in
TNFα signaling via NF-κB and interferon- α/γ response (Fig. 1I). We also analyzed
GO and KEGG enrichment by GSEA (Fig. 1J, K, respectively). GO enrichment indicated
that the tumor necrosis factor superfamily (TNFSF) cytokine production pathway was
enriched in low expression OSBPL3, while regulation of myeloid leukocyte mediated
immunity pathway was enriched in low expression TNRC6C (Fig. 1J). In the transcription
factors (TFs) analysis, we found these core genes were regulated by multiple TFs sharing
common mechanisms. These TFs were enriched by cumulative recovery curve, motif-TF
annotation, and selection of key genes (Fig. 1L, M). The motif with the highest normalized
enrichment score (NES: 8.79) was cisbp_M2425, and OSBPL3 was enriched in it (Fig. 1L).
The whole set of significant motifs (NES > 3) and corresponding TFs that were enriched
for either one or both of the two core genes are listed in Supplementary File 2. Lastly,
we performed qPCR to quantify the relative transcription levels of hub genes in SLE
and healthy controls (HCs). As shown in Figure 1N, OSBPL3 and TNRC6C transcription
levels were significantly decreased in SLE patients compared with HCs. To further
validate protein translation levels, we performed the western blotting of the PBMC
from SLE patients and HCs (each n = 6). We observed that the expression levels of
OSBPL3 and TNRC6C were greatly reduced in SLE patients (Fig. 1O).
Figure 1
Analysis of the study results. (
A, B
) Analysis of dataset GSE50772. (A) The volcano plot illustrates the DEGs between
control and SLE patients, including 3250 upregulated and 3567 downregulated genes.
(B) The heatmap clustering the expression of the top 10 upregulated or downregulated
DEGs among each sample. (
C
) The heatmap showing the DMPs between control and SLE patients, including 150 hypermethylated
probes (61 genes) and 88 hypomethylated probes (84 genes). (D
–
F) The WGCNA network analysis of 81 samples in dataset GSE50772. (D) Dendrogram of
co-expression modules (n = 18) identified by WGCNA, including black (n = 899), blue
(n = 1,858), cyan (n = 245), dark green (n = 177), dark orange (n = 126), dark red
(n = 307), green-yellow (n = 285), grey (n = 8,787), grey 60 (n = 477), light cyan
(n = 237), light green (n = 2,461), light yellow (n = 219), magenta (n = 564), orange
(n = 353), pink (n = 501), red (n = 702), tan (n = 280), and turquoise (n = 1,268)
modules. Different color branches of the cluster tree represent different modules,
and the color bands below the dendrogram show the cluster membership according to
different clustering methods. (E) Module–trait correlation heatmap. The color scale
on the right shows correlations from −1 (red) to 1 (blue). (F) A scatter plot showing
the correlation analysis between the gene significance score and module membership
in the light green module. (G) Two key genes identified by intersecting the light
green module with DEGs-hypermethylated genes. Relative expression of them in SLE and
control samples in the dataset was shown. (H–K) Functional analysis of the core genes.
(H) The correlation between OSBPL3 and TNRC6C and immune cell infiltration. (I) The
pathway enrichment analysis in high- or low-expression of OSBPL3 and TNRC6C in SLE
patients. (J) The enrichment of GO in high- or low-expression of OSBPL3 and TNRC6C
by GSEA analysis. (K) The enrichment of KEGG pathways in high- or low-expression of
OSBPL3 and TNRC6C by GSEA analysis. (L, M) Motif enrichment analysis of regulatory
regions in OSBPL3 and TNRC6C in SLE patients. (L) Analysis of global mean and SD estimation
as well as the recovery curves of motifs with high NES. The selection of significant
motifs is done based on NES, which is calculated for each motif based on the AUC distribution
of all the motifs for the gene set [(x-mean)/SD]. The red line is the mean of the
recovery curves of each motif. The green line is the mean value with standard deviation.
The blue line is the recovery curve of the indicated motif. (M) AUC histogram of hub
genes. (N, O) Relative transcription and translation levels of two hub genes. (N)
Relative transcription levels of OSBPL3 and TNRC6C in SLE patients (n = 22) or healthy
controls (n = 23) (both P = 0.0118). (O) Protein levels of OSBPL3 and TNRC6C in SLE
patients (n = 6) and healthy controls (n = 6). AUC, area under the curve; DC, dendritic
cell; GO, gene ontology; GSVA, gene set variation analysis; H, healthy control; HExp,
high expression; KEGG, Kyoto Encyclopedia of Genes and Genomes; LExp, low expression;
NK, natural killer; NS, no significant; NSE, normalized enrichment score; S, SLE patient;
SD, standard deviation; SLE, systemic lupus erythematosus. ∗P < 0.05. ∗∗P < 0.01.
Fig. 1
To date, distinctive transcriptional signatures have been identified in SLE, showing
the importance of how key genes play a role in its pathogenesis. OSBPL3 belongs to
the oxysterol-binding protein family and functions in cell adhesion, the actin cytoskeleton,
cellular lipid metabolism, vesicle transport, and cell signaling.
1
TNRC6C belongs to the trinucleotide repeat containing the 6/GW182 family and plays
important roles in RNA-mediated gene silencing by micro-RNAs.
2
In our analysis, they were downregulated in SLE patients, possibly due to hypermethylation
in their promoter region, which is consistent with the regulatory mechanism reported
in other diseases.
2
We also showed that downregulated OSBPL3 was associated with interleukin (IL)-8,
and TNFSF cytokine production and signaling, while downregulated TNRC6C was associated
with TNFα signaling via NF-κB. IL-8 is an important proinflammatory factor that induces
neutrophil activation, chemotaxis, and neutrophil extracellular trap (NET) formation.
3
NETs are one of the sources of self-antigens that destroy immune tolerance.
4
TNFSF ligand-receptors signaling pathways have been demonstrated to be active in
inflammatory and autoimmune disease.
5
Taken together, our findings imply that decreased OSBPL3 and TNRC6C might be involved
in the pro-inflammatory process in SLE. By analyzing immune cell infiltration, we
found both OSBPL3 and TNRC6C were inversely correlated with monocytes and neutrophils.
OSBPL3 was also inversely associated with activated dendritic cells, while TNRC6C
was inversely associated with plasma cells and memory B cells. Dendritic cells are
crucial in regulating peripheral tolerance to self-antigens in SLE and could induce
B cell differentiation, which ultimately leads to the production of pathogenic autoantibodies.
The interaction between the two hub genes and immune cells in the disease pathogenesis
should be highlighted in further research. In the TFs analysis, we identified TF regulatory
network associated with OSBPL3 and TNRC6C from publicly available SLE datasets. We
found that cisbp_M2425 was the most significantly enriched motif in OSBPL3, suggesting
a potential binding site of TFs.
In conclusion, based on bioinformatic data analysis, two novel hypermethylated, lowly
expressed hub genes (OSBPL3 and TNRC6C) were first identified and confirmed in SLE
patients. They may be involved in the pathophysiologic mechanism of SLE through aberrant
cell adhesion, induction of pro-inflammatory cytokines, and interaction with imbalanced
immune cells. Our study provides a new methodology to study the molecular mechanisms
of SLE from both epigenome and transcriptome. Further studies are warranted to validate
the hypothesis as well as the predicted TFs by regulatory network analysis.
Ethics declaration
This study was approved by the Ethical Committee of Sichuan Provincial People's Hospital.
All subjects signed informed consent.
Conflict of interests
The authors declare that they have no competing interests.
Funding
This research was supported by the
10.13039/501100001809
National Natural Science Foundation of China
(No. 81802504, 81872207), Sichuan Science and Technology Bureau, China (No. 2019YFS0439,
2020JDJQ0067, 2020JDRC0118, 2021YJ0564, and 2022YFH0005), the Science and Technology
Innovation Project of Chengdu, China (No. 2021-YF05-00225-SN), and a Sichuan Medical
Association grant (China) (No. Q19037).
Consent for publication
All authors signed a consent for publication form.
Availability of data and materials
The datasets analyzed during the current study are available in the GEO repository,
GSE50772: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50772.
GSE59250: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59250.
Related collections
Most cited references5
- Record: found
- Abstract: not found
- Article: not found
Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils.
M Baggiolini, A. Walz, S Kunkel (1989)
- Record: found
- Abstract: found
- Article: not found
TNF superfamily in inflammatory disease: translating basic insights.
Michael Croft, Wei Duan, Heonsik Choi … (2012)