- Record: found
- Abstract: found
- Article: not found
Development and evaluation of robust molecular markers linked to disease resistance in tomato for distinctness, uniformity and stability testing
Read this article at
Abstract
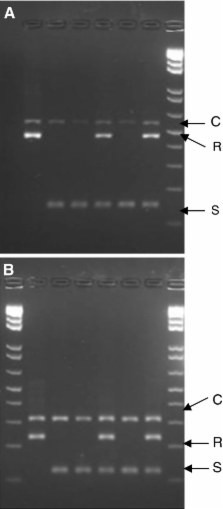
Molecular markers linked to phenotypically important traits are of great interest especially when traits are difficult and/or costly to be observed. In tomato where a strong focus on resistance breeding has led to the introgression of several resistance genes, resistance traits have become important characteristics in distinctness, uniformity and stability (DUS) testing for Plant Breeders Rights (PBR) applications. Evaluation of disease traits in biological assays is not always straightforward because assays are often influenced by environmental factors, and difficulties in scoring exist. In this study, we describe the development and/or evaluation of molecular marker assays for the Verticillium genes Ve1 and Ve2, the tomato mosaic virus Tm1 (linked marker), the tomato mosaic virus Tm2 and Tm2 2 genes, the Meloidogyne incognita Mi1-2 gene, the Fusarium I (linked marker) and I2 loci, which are obligatory traits in PBR testing. The marker assays were evaluated for their robustness in a ring test and then evaluated in a set of varieties. Although in general, results between biological assays and marker assays gave highly correlated results, marker assays showed an advantage over biological tests in that the results were clearer, i.e., homozygote/heterozygote presence of the resistance gene can be detected and heterogeneity in seed lots can be identified readily. Within the UPOV framework for granting of PBR, the markers have the potential to fulfil the requirements needed for implementation in DUS testing of candidate varieties and could complement or may be an alternative to the pathogenesis tests that are carried out at present.
Related collections
Most cited references18
- Record: found
- Abstract: not found
- Article: not found
An efficient procedure for genotyping single nucleotide polymorphisms.
- Record: found
- Abstract: found
- Article: not found
The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond.
- Record: found
- Abstract: found
- Article: not found
