- Record: found
- Abstract: found
- Article: found
Science for the Next Century: Deep Phenotyping
Read this article at
Abstract
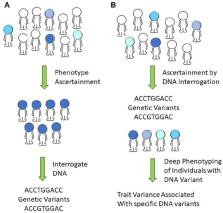
Our ability to unravel the mysteries of human health and disease have changed dramatically over the past 2 decades. Decoding health and disease has been facilitated by the recent availability of high-throughput genomics and multi-omics analyses and the companion tools of advanced informatics and computational science. Understanding of the human genome and its influence on phenotype continues to advance through genotyping large populations and using “light phenotyping” approaches in combination with smaller subsets of the population being evaluated using “deep phenotyping” approaches. Using our capability to integrate and jointly analyze genomic data with other multi-omic data, the knowledge of genotype-phenotype relationships and associated genetic pathways and functions is being advanced. Understanding genotype-phenotype relationships that discriminate human health from disease is speculated to facilitate predictive, precision health care and change modes of health care delivery. The American Association for Dental Research Fall Focused Symposium assembled experts to discuss how studies of genotype-phenotype relationships are illuminating the pathophysiology of craniofacial diseases and developmental biology. Although the breadth of the topic did not allow all areas of dental, oral, and craniofacial research to be addressed (e.g., cancer), the importance and power of integrating genomic, phenomic, and other -omic data are illustrated using a variety of examples. The 8 Fall Focused talks presented different methodological approaches for ascertaining study populations and evaluating population variance and phenotyping approaches. These advances are reviewed in this summary.
Related collections
Most cited references34

- Record: found
- Abstract: found
- Article: found
The UK Biobank resource with deep phenotyping and genomic data
- Record: found
- Abstract: found
- Article: not found
The sequence of the human genome.
- Record: found
- Abstract: found
- Article: not found