- Record: found
- Abstract: found
- Article: found
Hidden syndinian and perkinsid infections in dinoflagellate hosts revealed by single-cell transcriptomics
Read this article at
Abstract
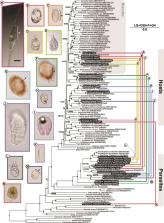
Free-living core dinoflagellates are commonly infected by members of two parasitic clades that are themselves closely related to dinoflagellates, the marine alveolates and perkinsids. These parasites are abundant and ecologically important, but most species have been difficult to observe directly or cultivate, so our knowledge of them is usually restricted to environmental 18S rRNA gene sequences, as genome-scale molecular data are not available for most species. Here, we report the finding of several of these parasites infecting free-living dinoflagellates. Of the 14 infected host cells collected, only five were identified as containing parasites via light microscopy at the time of collection. Single-cell transcriptome sequencing yielded relatively high transcriptomic coverage for parasites as well as their hosts. Host and parasite homologs were distinguished phylogenetically, allowing us to infer a robust phylogenomic tree based on 192 genes. The tree showed one parasite belongs to an undescribed lineage that is sister to perkinsids, whereas the remainder are members of the syndinian clade within the marine alveolates. Close relatives of all these parasites have been observed in 18S rRNA gene surveys, but until now none had been linked to a specific host. These findings illustrate the efficacy of single-cell isolation and transcriptome sequencing as strategies for gaining deeper insights into the evolutionary history and host relationships of hidden single-celled parasites.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean.
- Record: found
- Abstract: found
- Article: not found
Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton.
- Record: found
- Abstract: found
- Article: not found
Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.