- Record: found
- Abstract: found
- Article: found
The torso-like gene functions to maintain the structure of the vitelline membrane in Nasonia vitripennis, implying its co-option into Drosophila axis formation
Read this article at
ABSTRACT
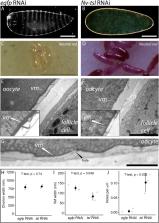
Axis specification is a fundamental developmental process. Despite this, the mechanisms by which it is controlled across insect taxa are strikingly different. An excellent example of this is terminal patterning, which in Diptera such as Drosophila melanogaster occurs via the localized activation of the receptor tyrosine kinase Torso. In Hymenoptera, however, the same process appears to be achieved via localized mRNA . How these mechanisms evolved and what they evolved from remains largely unexplored. Here, we show that torso-like, known for its role in Drosophila terminal patterning, is instead required for the integrity of the vitelline membrane in the hymenopteran wasp Nasonia vitripennis. We find that other genes known to be involved in Drosophila terminal patterning, such as torso and Ptth, also do not function in Nasonia embryonic development. These findings extended to orthologues of Drosophila vitelline membrane proteins known to play a role in localizing Torso-like in Drosophila; in Nasonia these are instead required for dorso–ventral patterning, gastrulation and potentially terminal patterning. Our data underscore the importance of the vitelline membrane in insect development, and implies phenotypes caused by knockdown of torso-like must be interpreted in light of its function in the vitelline membrane. In addition, our data imply that the signalling components of the Drosophila terminal patterning systems were co-opted from roles in regulating moulting, and co-option into terminal patterning involved the evolution of a novel interaction with the vitelline membrane protein Torso-like.
This article has an associated First Person interview with the first author of the paper.
Abstract
Summary: In the parasitic wasp Nasonia, Tsl, a key component of the process that defines the termini of the embryo of Drosophila, has a function in the structure of the vitelline membrane.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases.
- Record: found
- Abstract: found
- Article: not found
Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects.
- Record: found
- Abstract: found
- Article: not found