- Record: found
- Abstract: found
- Article: found
Predictive evolution of metabolic phenotypes using model‐designed environments
Read this article at
Abstract
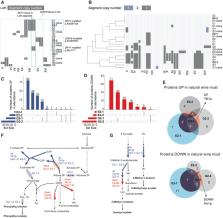
Adaptive evolution under controlled laboratory conditions has been highly effective in selecting organisms with beneficial phenotypes such as stress tolerance. The evolution route is particularly attractive when the organisms are either difficult to engineer or the genetic basis of the phenotype is complex. However, many desired traits, like metabolite secretion, have been inaccessible to adaptive selection due to their trade‐off with cell growth. Here, we utilize genome‐scale metabolic models to design nutrient environments for selecting lineages with enhanced metabolite secretion. To overcome the growth‐secretion trade‐off, we identify environments wherein growth becomes correlated with a secondary trait termed tacking trait. The latter is selected to be coupled with the desired trait in the application environment where the trait manifestation is required. Thus, adaptive evolution in the model‐designed selection environment and subsequent return to the application environment is predicted to enhance the desired trait. We experimentally validate this strategy by evolving Saccharomyces cerevisiae for increased secretion of aroma compounds, and confirm the predicted flux‐rerouting using genomic, transcriptomic, and proteomic analyses. Overall, model‐designed selection environments open new opportunities for predictive evolution.
Abstract
Related collections
Most cited references100

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.

- Record: found
- Abstract: found
- Article: found
