- Record: found
- Abstract: found
- Article: found
Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons
Read this article at
Abstract
Background
Polyploidy is a pervasive evolutionary feature of all flowering plants and some animals, leading to genetic and epigenetic changes that affect gene expression and morphology. DNA methylation changes can produce meiotically stable epialleles, which are transmissible through selection and breeding. However, the relationship between DNA methylation and polyploid plant domestication remains elusive.
Results
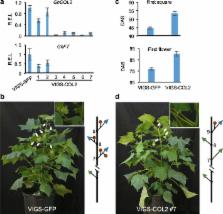
We report comprehensive epigenomic and functional analyses, including ~12 million differentially methylated cytosines in domesticated allotetraploid cottons and their tetraploid and diploid relatives. Methylated genes evolve faster than unmethylated genes; DNA methylation changes between homoeologous loci are associated with homoeolog-expression bias in the allotetraploids. Significantly, methylation changes induced in the interspecific hybrids are largely maintained in the allotetraploids. Among 519 differentially methylated genes identified between wild and cultivated cottons, some contribute to domestication traits, including flowering time and seed dormancy. CONSTANS ( CO) and CO- LIKE ( COL) genes regulate photoperiodicity in Arabidopsis. COL2 is an epiallele in allotetraploid cottons. COL2A is hypermethylated and silenced, while COL2D is repressed in wild cottons but highly expressed due to methylation loss in all domesticated cottons tested. Inhibiting DNA methylation activates COL2 expression, and repressing COL2 in cultivated cotton delays flowering.
Conclusions
We uncover epigenomic signatures of domestication traits during cotton evolution. Demethylation of COL2 increases its expression, inducing photoperiodic flowering, which could have contributed to the suitability of cotton for cultivation worldwide. These resources should facilitate epigenetic engineering, breeding, and improvement of polyploid crops.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening.
- Record: found
- Abstract: not found
- Article: not found
Targeted mutation of the DNA methyltransferase gene results in embryonic lethality
- Record: found
- Abstract: found
- Article: not found