- Record: found
- Abstract: found
- Article: found
Streamlined selection of cancer antigens for vaccine development through integrative multi-omics and high-content cell imaging
Read this article at
Abstract
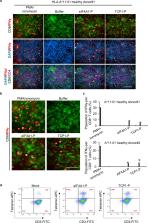
Identification of tumor antigens that induce cytotoxic T lymphocytes (CTLs) is crucial for cancer-vaccine development. Despite their predictive ability, current algorithmic approaches and human leukocyte antigen (HLA)-peptidomic analysis allow limited selectivity. Here, we optimized a method to rapidly screen and identify highly immunogenic epitopes that trigger CTL responses. We used a combined application of this method involving immune-specific signature analysis and HLA-associated peptidomics using samples from six patients with triple-negative breast cancer (TNBC) in order to select immunogenic HLA epitopes for in vitro testing. Additionally, we applied high-throughput imaging at the single-cell level in order to confirm the immunoreactivity of the selected peptides. The results indicated that this method enabled identification of promising CTL peptides capable of inducing antitumor immunity. This platform combining high-resolution computational analysis, HLA-peptidomics, and high-throughput immunogenicity testing allowed rapid and robust identification of highly immunogenic epitopes and represents a powerful technique for cancer-vaccine development.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy.
- Record: found
- Abstract: found
- Article: not found
Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells.
- Record: found
- Abstract: found
- Article: not found