- Record: found
- Abstract: found
- Article: found
Identification of cuproptosis‐related lncRNAs for prognosis and immunotherapy in glioma
Read this article at
Abstract
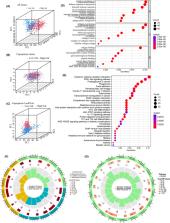
Glioma is a highly invasive primary brain tumour, making it challenging to accurately predict prognosis for glioma patients. Cuproptosis is a recently discovered cell death attracting significant attention in the tumour field. Whether cuproptosis‐related genes have prognostic predictive value has not been clarified. In this study, uni‐/multi‐variate Cox and Lasso regression analyses were applied to construct a risk model based on cuproptosis‐related lncRNAs using TCGA and CGGA cohorts. A nomogram was constructed to quantify individual risk, including clinical and genic characteristics and risk. GO and KEGG analyses were used to define functional enrichment of DEGs. Tumour mutation burden (TMB) and immune checkpoint analyses were performed to evaluate potential responses to ICI therapy. Ten prognostic lncRNAs were obtained from Cox regression. Based on the median risk score, patients were divided into high‐ and low‐risk groups. Either for grade 2–3 or for grade 4, glioma patients with high‐risk exhibited significant poorer prognoses. The risk was an independent risk factor associated with overall survival. The high‐risk group was functionally associated with immune responses and cancer‐related pathways. The high‐risk group was associated with higher TMB scores. The expression levels of many immune checkpoints in the high‐risk group were significantly higher than those in the low‐risk group. Differentiated immune pathways were primarily enriched in the IFN response, immune checkpoint and T‐cell co‐stimulation pathways. In conclusion, we established a risk model based on cuproptosis‐related lncRNAs showing excellent prognostic prediction ability but also indicating the immuno‐microenvironment status of glioma.
Related collections
Most cited references41

- Record: found
- Abstract: found
- Article: found
limma powers differential expression analyses for RNA-sequencing and microarray studies

- Record: found
- Abstract: found
- Article: found
Maftools: efficient and comprehensive analysis of somatic variants in cancer
- Record: found
- Abstract: found
- Article: not found