- Record: found
- Abstract: found
- Article: found
Minimal Residual Disease Detection in Acute Lymphoblastic Leukemia
Read this article at
Abstract
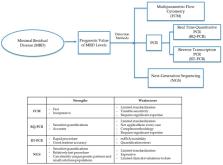
Minimal residual disease (MRD) refers to a chemotherapy/radiotherapy-surviving leukemia cell population that gives rise to relapse of the disease. The detection of MRD is critical for predicting the outcome and for selecting the intensity of further treatment strategies. The development of various new diagnostic platforms, including next-generation sequencing (NGS), has introduced significant advances in the sensitivity of MRD diagnostics. Here, we review current methods to diagnose MRD through phenotypic marker patterns or differential gene patterns through analysis by flow cytometry (FCM), polymerase chain reaction (PCR), real-time quantitative polymerase chain reaction (RQ-PCR), reverse transcription polymerase chain reaction (RT-PCR) or NGS. Future advances in clinical procedures will be molded by practical feasibility and patient needs regarding greater diagnostic sensitivity.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
High-throughput sequencing technologies.
- Record: found
- Abstract: found
- Article: found
Acute lymphoblastic leukemia: a comprehensive review and 2017 update

- Record: found
- Abstract: found
- Article: found