- Record: found
- Abstract: found
- Article: found
Altered microbiota, fecal lactate, and fecal bile acids in dogs with gastrointestinal disease
Read this article at
Abstract
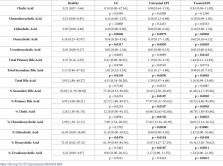
The intestinal microbiota plays an important role in health and disease and produces, through fermentative reactions, several metabolic products, such as lactate, that can affect the host. The microbiota also interacts with and metabolizes compounds produced by the host, such as primary bile acids. Lactate and bile acids (BA) are of particular interest in gastrointestinal diseases because they have been associated with metabolic acidosis and bile acid diarrhea, respectively. The objectives of this study were to validate an enzymatic assay to quantify D-, L-, and total lactate in canine feces, and to characterize fecal lactate and BA concentrations as well as bacterial abundances in healthy dogs and dogs with gastrointestinal diseases. Fecal samples were collected from 34 healthy dogs, 15 dogs with chronic enteropathy (CE), and 36 dogs with exocrine pancreatic insufficiency (EPI). Lactate was quantified with an enzymatic assay, BA with gas chromatography-mass spectrometry, and 11 bacterial groups with qPCR. A fecal lactate reference interval was established from 34 healthy dogs and was 0.7–1.4 mM, 0.3–6.0 mM, and 1.0–7.0 mM for D-, L-, and total lactate, respectively. The assay to measure D-, L-, and total lactate in canine fecal samples was linear, accurate, precise, and reproducible. Significant increases in fecal lactate and decreases in secondary BA concentrations were observed in dogs with CE and dogs with EPI. Dogs with EPI had an increased abundance of Escherichia coli, Lactobacillus, and Bifidobacterium; a decreased abundance of Fusobacterium and Clostridium hiranonis; and a higher Dysbiosis Index when compared to healthy dogs. Further studies are necessary to determine the clinical utility of lactate and BA quantification in canine feces. These metabolites suggest functional alterations of intestinal dysbiosis and may become promising targets for further elucidating the role of the microbiota in health and disease.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases.
- Record: found
- Abstract: found
- Article: not found
Utilization of nutrients by isolated epithelial cells of the rat colon.
- Record: found
- Abstract: found
- Article: not found